An Efficient, Scalable, and Exact Representation of High-Dimensional Color Information Enabled Using de Bruijn Graph Search
- PMID: 32176522
- PMCID: PMC7185321
- DOI: 10.1089/cmb.2019.0322
An Efficient, Scalable, and Exact Representation of High-Dimensional Color Information Enabled Using de Bruijn Graph Search
Abstract
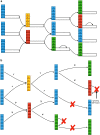
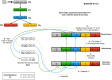
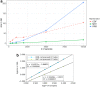
The colored de Bruijn graph (cdbg) and its variants have become an important combinatorial structure used in numerous areas in genomics, such as population-level variation detection in metagenomic samples, large-scale sequence search, and cdbg-based reference sequence indices. As samples or genomes are added to the cdbg, the color information comes to dominate the space required to represent this data structure. In this article, we show how to represent the color information efficiently by adopting a hierarchical encoding that exploits correlations among color classes-patterns of color occurrence-present in the de Bruijn graph (dbg). A major challenge in deriving an efficient encoding of the color information that takes advantage of such correlations is determining which color classes are close to each other in the high-dimensional space of possible color patterns. We demonstrate that the dbg itself can be used as an efficient mechanism to search for approximate nearest neighbors in this space. While our approach reduces the encoding size of the color information even for relatively small cdbgs (hundreds of experiments), the gains are particularly consequential as the number of potential colors (i.e., samples or references) grows into thousands. We apply this encoding in the context of two different applications; the implicit cdbg used for a large-scale sequence search index, Mantis, as well as the encoding of color information used in population-level variation detection by tools such as Vari and Rainbowfish. Our results show significant improvements in the overall size and scalability of representation of the color information. In our experiment on 10,000 samples, we achieved >11 × better compression compared to Ramen, Ramen, Rao (RRR).
Keywords: RNA-sequence search; compression schemes; de bruijn graph; proximate membership query.
Conflict of interest statement
The authors declare they have no competing financial interests.
Figures

References
-
- Alipanahi B., Kuhnle A., and Boucher C.. 2018a. Recoloring the colored de Bruijn graph, 1–11. In International Symposium on String Processing and Information Retrieval. Springer, Cham
-
- Alipanahi B., Muggli M. D., Jundi M., et al. 2018b. Resistome SNP calling via read colored de Bruijn graphs. bioRxiv, 156174 Technical Report 312, Operation Research, NC state university, Waden, Germany
-
- Almodaresi F., Pandey P., and Patro R.. 2017. Rainbowfish: A succinct colored de Bruijn graph representation. In LIPIcs-Leibniz International Proceedings in Informatics, volume 88 Schloss Dagstuhl-Leibniz-Zentrum fuer Informatik. Oxford University Press, Oxford, England
-
- Althaus E., Funke S., Har-Peled S., et al. 2005. Approximating k-hop minimum-spanning trees. Oper. Res. Lett. 33, 115–120
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

