Checkpoint Responses to DNA Double-Strand Breaks
- PMID: 32176524
- PMCID: PMC7311309
- DOI: 10.1146/annurev-biochem-011520-104722
Checkpoint Responses to DNA Double-Strand Breaks
Abstract
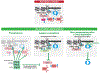
Cells confront DNA damage in every cell cycle. Among the most deleterious types of DNA damage are DNA double-strand breaks (DSBs), which can cause cell lethality if unrepaired or cancers if improperly repaired. In response to DNA DSBs, cells activate a complex DNA damage checkpoint (DDC) response that arrests the cell cycle, reprograms gene expression, and mobilizes DNA repair factors to prevent the inheritance of unrepaired and broken chromosomes. Here we examine the DDC, induced by DNA DSBs, in the budding yeast model system and in mammals.
Keywords: DNA double-strand break; DNA repair; cell cycle; checkpoint; kinases.
Figures

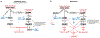


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

