Origin, genomic diversity and microevolution of the Clostridium difficile B1/NAP1/RT027/ST01 strain in Costa Rica, Chile, Honduras and Mexico
- PMID: 32176604
- PMCID: PMC7371124
- DOI: 10.1099/mgen.0.000355
Origin, genomic diversity and microevolution of the Clostridium difficile B1/NAP1/RT027/ST01 strain in Costa Rica, Chile, Honduras and Mexico
Abstract
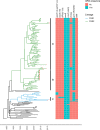
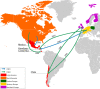
Clostridium difficile B1/NAP1/RT027/ST01 has been responsible for outbreaks of antibiotic-associated diarrhoea in clinical settings worldwide and is associated with severe disease presentations and increased mortality rates. Two fluoroquinolone-resistant (FQR) lineages of the epidemic B1/NAP1/RT027/ST01 strain emerged in the USA in the early 1990s and disseminated trans continentally (FQR1 and FQR2). However, it is unclear when and from where they entered Latin America (LA) and whether isolates from LA exhibit unique genomic features when compared to B1/NAP1/RT027/ST01 isolates from other regions of the world. To answer the first issue we compared whole-genome sequences (WGS) of 25 clinical isolates typed as NAP1, RT027 or ST01 in Costa Rica (n=16), Chile (n=5), Honduras (n=3) and Mexico (n=1) to WGS of 129 global isolates from the same genotype using Bayesian phylogenomics. The second question was addressed through a detailed analysis of the number and type of mutations of the LA isolates and their mobile resistome. All but two B1/NAP1/RT027/ST01 isolates from LA belong to the FQR2 lineage (n=23, 92 %), confirming its widespread distribution. As indicated by analysis of a dataset composed of 154 WGS, the B1/NAP1/RT027/ST01 strain was introduced into the four LA countries analysed between 1998 and 2005 from North America (twice) and Europe (at least four times). These events occurred soon after the emergence of the FQR lineages and more than one decade before the first report of the detection of the B1/NAP1/RT027/ST01 in LA. A total of 552 SNPs were identified across all genomes examined (3.8-4.3 Mb) in pairwise comparisons to the R20291 reference genome. Moreover, pairwise SNP distances were among the smallest distances determined in this species so far (0 to 55). Despite this high level of genomic conservation, 39 unique SNPs (7 %) in genes that play roles in the infection process (i.e. slpA) or antibiotic resistance (i.e. rpoB, fusA) distinguished the LA isolates. In addition, isolates from Chile, Honduras and Mexico had twice as many antibiotic resistance genes (ARGs, n=4) than related isolates from other regions. Their unique set of ARGs includes a cfr-like gene and tetM, which were found as part of putative mobile genetic elements whose sequences resemble undescribed integrative and conjugative elements. These results show multiple, independent introductions of B1/NAP1/RT027/ST01 isolates from the FQR1 and FQR2 lineages from different geographical sources into LA and a rather rapid accumulation of distinct mutations and acquired ARG by the LA isolates.
Keywords: B1/NAP1/RT027/ST01 strain; Clostridium difficile; Latin America; MGEs; phylogenomics.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

