The use of mixture density networks in the emulation of complex epidemiological individual-based models
- PMID: 32176687
- PMCID: PMC7098654
- DOI: 10.1371/journal.pcbi.1006869
The use of mixture density networks in the emulation of complex epidemiological individual-based models
Abstract
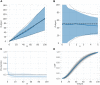
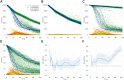
Complex, highly-computational, individual-based models are abundant in epidemiology. For epidemics such as macro-parasitic diseases, detailed modelling of human behaviour and pathogen life-cycle are required in order to produce accurate results. This can often lead to models that are computationally-expensive to analyse and perform model fitting, and often require many simulation runs in order to build up sufficient statistics. Emulation can provide a more computationally-efficient output of the individual-based model, by approximating it using a statistical model. Previous work has used Gaussian processes (GPs) in order to achieve this, but these can not deal with multi-modal, heavy-tailed, or discrete distributions. Here, we introduce the concept of a mixture density network (MDN) in its application in the emulation of epidemiological models. MDNs incorporate both a mixture model and a neural network to provide a flexible tool for emulating a variety of models and outputs. We develop an MDN emulation methodology and demonstrate its use on a number of simple models incorporating both normal, gamma and beta distribution outputs. We then explore its use on the stochastic SIR model to predict the final size distribution and infection dynamics. MDNs have the potential to faithfully reproduce multiple outputs of an individual-based model and allow for rapid analysis from a range of users. As such, an open-access library of the method has been released alongside this manuscript.
Conflict of interest statement
The authors MI and QC declare that they are members of the data science consultancy Scai Analytics Ltd. All other authors declare that they have no competing interests.
Figures



Similar articles
-
Gaussian process approximations for fast inference from infectious disease data.Math Biosci. 2018 Jul;301:111-120. doi: 10.1016/j.mbs.2018.02.003. Epub 2018 Feb 20. Math Biosci. 2018. PMID: 29471011
-
Continuous and discrete SIR-models with spatial distributions.J Math Biol. 2017 Jun;74(7):1709-1727. doi: 10.1007/s00285-016-1071-8. Epub 2016 Oct 28. J Math Biol. 2017. PMID: 27796478
-
Dynamics of stochastic epidemics on heterogeneous networks.J Math Biol. 2014 Jun;68(7):1583-605. doi: 10.1007/s00285-013-0679-1. Epub 2013 Apr 30. J Math Biol. 2014. PMID: 23633042
-
Modeling epidemics: A primer and Numerus Model Builder implementation.Epidemics. 2018 Dec;25:9-19. doi: 10.1016/j.epidem.2018.06.001. Epub 2018 Jul 13. Epidemics. 2018. PMID: 30017895 Review.
-
Epidemionics: from the host-host interactions to the systematic analysis of the emergent macroscopic dynamics of epidemic networks.Virulence. 2010 Jul-Aug;1(4):338-49. doi: 10.4161/viru.1.4.12196. Virulence. 2010. PMID: 21178467 Review.
Cited by
-
Using mixture density networks to emulate a stochastic within-host model of Francisella tularensis infection.PLoS Comput Biol. 2023 Dec 20;19(12):e1011266. doi: 10.1371/journal.pcbi.1011266. eCollection 2023 Dec. PLoS Comput Biol. 2023. PMID: 38117811 Free PMC article.
-
Parametric seasonal-trend autoregressive neural network for long-term crop price forecasting.PLoS One. 2024 Sep 26;19(9):e0311199. doi: 10.1371/journal.pone.0311199. eCollection 2024. PLoS One. 2024. PMID: 39325794 Free PMC article.
-
Optimizing COVID-19 vaccine distribution across the United States using deterministic and stochastic recurrent neural networks.PLoS One. 2021 Jul 6;16(7):e0253925. doi: 10.1371/journal.pone.0253925. eCollection 2021. PLoS One. 2021. PMID: 34228740 Free PMC article.
-
Approximating solutions of the Chemical Master equation using neural networks.iScience. 2022 Aug 27;25(9):105010. doi: 10.1016/j.isci.2022.105010. eCollection 2022 Sep 16. iScience. 2022. PMID: 36117994 Free PMC article.
-
Distilling dynamical knowledge from stochastic reaction networks.Proc Natl Acad Sci U S A. 2024 Apr 2;121(14):e2317422121. doi: 10.1073/pnas.2317422121. Epub 2024 Mar 26. Proc Natl Acad Sci U S A. 2024. PMID: 38530895 Free PMC article.
References
-
- Keeling MJ, Rohani P. Modeling infectious diseases in humans and animals. Princeton University Press; 2011.
-
- May RM. Togetherness among schistosomes: its effects on the dynamics of the infection. Mathematical Biosciences. 1977;35(3-4):301–343. 10.1016/0025-5564(77)90030-X - DOI
-
- Irvine MA, Reimer LJ, Njenga SM, Gunawardena S, Kelly-Hope L, Bockarie M, et al. Modelling strategies to break transmission of lymphatic filariasis-aggregation, adherence and vector competence greatly alter elimination. Parasites & Vectors. 2015;8(1):547 10.1186/s13071-015-1152-3 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

