The liver as an immunological barrier redefined by single-cell analysis
- PMID: 32176810
- PMCID: PMC7218664
- DOI: 10.1111/imm.13193
The liver as an immunological barrier redefined by single-cell analysis
Abstract
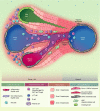
The liver is a front-line immune tissue that plays a major role in the detection, capture and clearance of pathogens and foreign antigens entering the bloodstream, especially from the gut. Our largest internal organ maintains this immune barrier in the face of constant exposure to external but harmless antigens through a highly specialized network of liver-adapted immune cells. Mapping the immune resident compartment in the liver has been challenging because it requires multimodal single-cell deep phenotyping approaches of often rare cell populations in difficult to access samples. We can now measure the RNA transcripts present in a single cell (scRNA-seq), which is revolutionizing the way we characterize cell types. scRNA-seq has been applied to the diverse array of immune cells present in murine and human livers in health and disease. Here, we summarize how emerging single-cell technologies have advanced or redefined our understanding of the immunological barrier provided by the liver.
Keywords: RNA-seq; immune barrier; liver; liver resident cells; single cells; transcriptomics.
© 2020 The Authors. Immunology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kubes P, Jenne C. Immune responses in the liver. Annu Rev Immunol 2018; 36:247–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

