The Increase of Simple Sequence Repeats during Diversification of Marchantiidae, An Early Land Plant Lineage, Leads to the First Known Expansion of Inverted Repeats in the Evolutionarily-Stable Structure of Liverwort Plastomes
- PMID: 32178248
- PMCID: PMC7140840
- DOI: 10.3390/genes11030299
The Increase of Simple Sequence Repeats during Diversification of Marchantiidae, An Early Land Plant Lineage, Leads to the First Known Expansion of Inverted Repeats in the Evolutionarily-Stable Structure of Liverwort Plastomes
Abstract
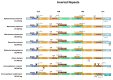
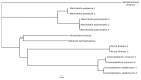
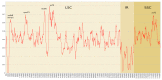
The chloroplast genomes of liverworts, an early land plant lineage, exhibit stable structure and gene content, however the known resources are very limited. The newly sequenced plastomes of Conocephalum, Riccia and Sphaerocarpos species revealed an increase of simple sequence repeats during the diversification of complex thalloid liverwort lineage. The presence of long TA motifs forced applying the long-read nanopore sequencing method for proper and dependable plastome assembly, since the length of dinucleotide repeats overcome the length of Illumina short reads. The accumulation of SSRs (simple sequence repeats) enabled the expansion of inverted repeats by the incorporation of rps12 and rps7 genes, which were part of large single copy (LSC) regions in the previously sequenced plastomes. The expansion of inverted repeat (IR) at the genus level is reported for the first time for non-flowering plants. Moreover, comparative analyses with remaining liverwort lineages revealed that the presence of SSR in plastomes is specific for simple thalloid species. Phylogenomic analysis resulted in trees confirming monophyly of Marchantiidae and partially congruent with previous studies, due to dataset-dependent results of Dumortiera-Reboulia relationships. Despite the lower evolutionary rate of Marchantiales plastomes, significant barcoding gap was detected, even for recently divergent holarctic Conocephalum species. The sliding window analyses revealed the presence of 18 optimal (500 bp long) barcodes that enable the molecular identification of all studied species.
Keywords: chloroplast genome; cpSSR; inverted repeats expansion; liverworts; nanopore sequencing; phylogeny; super-barcoding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Vitelli M., Vessella F., Cardoni S., Pollegioni P., Denk T., Grimm G.W., Simeone M.C. Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus Group Ilex, Fagaceae) Tree Genet. Genomes. 2017;13 doi: 10.1007/s11295-016-1086-8. - DOI
-
- Szczecińska M., Łazarski G., Bilska K., Sawicki J. The complete plastid genome and nuclear genome markers provide molecular evidence for the hybrid origin of pulsatilla × hackelii Pohl. Turk. J. Bot. 2017;41:329–337. doi: 10.3906/bot-1610-28. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

