LncRNA LEF1-AS1 silencing diminishes EZH2 expression to delay hepatocellular carcinoma development by impairing CEBPB-interaction with CDCA7
- PMID: 32178558
- PMCID: PMC7217375
- DOI: 10.1080/15384101.2020.1731052
LncRNA LEF1-AS1 silencing diminishes EZH2 expression to delay hepatocellular carcinoma development by impairing CEBPB-interaction with CDCA7
Abstract
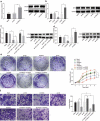
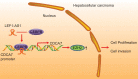
Hepatocellular carcinoma (HCC) is recognized for its high mortality rate worldwide. Based on intensive studies, long non-coding RNA (lncRNA) expression exerts significant effects on tumor suppression. Herein, we investigated the molecular mechanism of lymphoid enhancer-binding factor-1 antisense RNA 1 (LEF1-AS1) in HCC cells. Microarray-based gene expression analysis was adopted to predict and verify the differentially expressed genes in HCC, which predicted cell division cycle-associated 7 (CDCA7) and LEF1-AS1 to be highly expressed in HCC. The expression of LEF1-AS1, CDCA7, CCAAT/enhancer-binding protein beta (CEBPB) and enhancer of zeste homolog 2 (EZH2) was determined by means of reverse transcription quantitative polymerase chain reaction (RT-qPCR) and western blot analysis. LncMap was used to predict the lncRNA-transcription factor-gene interaction in HCC. ChIP, RIP assay and dual luciferase reporter gene assay were employed to verify the relationship between the transcription factor and gene. Silencing of LEF1-AS1 could downregulate CDCA7 expression through CEBPB. Overexpression of LEF1-AS1, EZH2 and CDCA7 promoted proliferation and invasion in HCC cells. LEF1-AS1 promoted CDCA7 expression to further upregulate EZH2. Tumor formation in nude mice was assessed to verify the experimental results. Silencing of LEF1-AS1 inhibited the growth of tumors in vivo. Collectively, silencing LEF1-AS1 inhibited the proliferation and invasion of HCC cells by down-regulating EZH2 through the CEBPB-CDCA7 signaling pathway, which provides scientific evidence for the treatment of HCC.Abbreviations: HCC: Hepatocellular carcinoma; lncRNA: long non-coding RNA; LEF1-AS1: lymphoid enhancer-binding factor-1 antisense RNA 1; EZH2: enhancer of zeste homolog 2; CDCA7: cell division cycle-associated 7; GEO: Gene Expression Omnibus; NC: negative control; oe: overexpressed; RT-qPCR: reverse transcription quantitative polymerase chain reaction; PBS: phosphate buffered saline; HRP: horseradish peroxidase; OD: optical density; RIP: Radioimmunoprecipitation; ChIP: Chromatin immunoprecipitation; WT: wild type.
Keywords: CDCA7; CEBPB; EZH2; LEF1-AS1; hepatocellular carcinoma; invasion; proliferation; transcription factor.
Figures




References
-
- Sherman M. Hepatocellular carcinoma: epidemiology, surveillance, and diagnosis. Semin Liver Dis. 2010. February;30(1):3–16. PubMed PMID: 20175029. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
