Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26
- PMID: 32178593
- PMCID: PMC7103712
- DOI: 10.1080/22221751.2020.1739565
Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26
Abstract
The recent outbreak of pneumonia-causing COVID-19 in China is an urgent global public health issue with an increase in mortality and morbidity. Here we report our modelled homo-trimer structure of COVID-19 spike glycoprotein in both closed (ligand-free) and open (ligand-bound) conformation, which is involved in host cell adhesion. We also predict the unique N- and O-linked glycosylation sites of spike glycoprotein that distinguish it from the SARS and underlines shielding and camouflage of COVID-19 from the host the defence system. Furthermore, our study also highlights the key finding that the S1 domain of COVID-19 spike glycoprotein potentially interacts with the human CD26, a key immunoregulatory factor for hijacking and virulence. These findings accentuate the unique features of COVID-19 and assist in the development of new therapeutics.
Keywords: CD26; Coronavirus; docking; glycosylation; spike glycoprotein.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

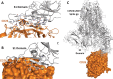
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
