High-throughput enrichment of temperature-sensitive argininosuccinate synthetase for two-stage citrulline production in E. coli
- PMID: 32179161
- PMCID: PMC7225747
- DOI: 10.1016/j.ymben.2020.03.004
High-throughput enrichment of temperature-sensitive argininosuccinate synthetase for two-stage citrulline production in E. coli
Abstract
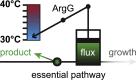
Controlling metabolism of engineered microbes is important to modulate cell growth and production during a bioprocess. For example, external parameters such as light, chemical inducers, or temperature can act on metabolism of production strains by changing the abundance or activity of enzymes. Here, we created temperature-sensitive variants of an essential enzyme in arginine biosynthesis of Escherichia coli (argininosuccinate synthetase, ArgG) and used them to dynamically control citrulline overproduction and growth of E. coli. We show a method for high-throughput enrichment of temperature-sensitive ArgG variants with a fluorescent TIMER protein and flow cytometry. With 90 of the thus derived ArgG variants, we complemented an ArgG deletion strain showing that 90% of the strains exhibit temperature-sensitive growth and 69% of the strains are auxotrophic for arginine at 42 °C and prototrophic at 30 °C. The best temperature-sensitive ArgG variant enabled precise and tunable control of cell growth by temperature changes. Expressing this variant in a feedback-dysregulated E. coli strain allowed us to realize a two-stage bioprocess: a 33 °C growth-phase for biomass accumulation and a 39 °C stationary-phase for citrulline production. With this two-stage strategy, we produced 3 g/L citrulline during 45 h cultivation in a 1-L bioreactor. These results show that temperature-sensitive enzymes can be created en masse and that they may function as metabolic valves in engineered bacteria.
Keywords: Citrulline overproduction; Flow cytometry; Single-cell growth; Temperature-sensitive enzymes; Two-stage bioprocess.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest None.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

