A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes
- PMID: 32179627
- PMCID: PMC7221254
- DOI: 10.1128/JB.00115-20
A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes
Abstract
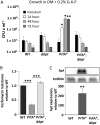
Listeria monocytogenes is a Gram-positive pathogen able to cause severe human infections. Its major virulence regulator is the transcriptional activator PrfA, a member of the Crp/Fnr family of transcriptional regulators. To establish a successful L. monocytogenes infection, the PrfA protein needs to be in an active conformation, either by binding the cognate inducer glutathione (GSH) or by possessing amino acid substitutions rendering the protein constitutively active (PrfA*). By a yet unknown mechanism, phosphotransferase system (PTS) sugars repress the activity of PrfA. We therefore took a transposon-based approach to identify the mechanism by which PTS sugars repress PrfA activity. For this, we screened a transposon mutant bank to identify clones able to grow in the presence of glucose-6-phosphate as the sole carbon source. Surprisingly, most of the isolated transposon mutants also carried amino acid substitutions in PrfA. In transposon-free strains, the PrfA amino acid substitution mutants displayed growth, virulence factor expression, infectivity, and DNA binding, agreeing with previously identified PrfA* mutants. Hence, the initial growth phenotype observed in the isolated clone was due to the amino acid substitution in PrfA and unrelated to the loci inactivated by the transposon mutant. Finally, we provide structural evidence for the existence of an intermediately activated PrfA state, which gives new insights into PrfA protein activation.IMPORTANCE The Gram-positive bacterium Listeria monocytogenes is a human pathogen affecting mainly the elderly, immunocompromised people, and pregnant women. It can lead to meningoencephalitis, septicemia, and abortion. The major virulence regulator in L. monocytogenes is the PrfA protein, a transcriptional activator. Using a growth-based selection strategy, we identified mutations in the PrfA protein leading to constitutively active virulence factor expression. We provide structural evidence for the existence of an intermediately activated PrfA state, which gives new insights into PrfA protein activation.
Keywords: ActA; LLO; Listeria monocytogenes; PrfA; PrfA*; crystal structure.
Copyright © 2020 Hansen et al.
Figures
Similar articles
-
Free Fatty Acids Interfere with the DNA Binding Activity of the Virulence Regulator PrfA of Listeria monocytogenes.J Bacteriol. 2020 Jul 9;202(15):e00156-20. doi: 10.1128/JB.00156-20. Print 2020 Jul 9. J Bacteriol. 2020. PMID: 32393522 Free PMC article.
-
Spontaneous Loss of Virulence in Natural Populations of Listeria monocytogenes.Infect Immun. 2017 Oct 18;85(11):e00541-17. doi: 10.1128/IAI.00541-17. Print 2017 Nov. Infect Immun. 2017. PMID: 28827366 Free PMC article.
-
The mutation G145S in PrfA, a key virulence regulator of Listeria monocytogenes, increases DNA-binding affinity by stabilizing the HTH motif.Mol Microbiol. 2005 Apr;56(2):433-46. doi: 10.1111/j.1365-2958.2005.04561.x. Mol Microbiol. 2005. PMID: 15813735
-
Cross Talk between SigB and PrfA in Listeria monocytogenes Facilitates Transitions between Extra- and Intracellular Environments.Microbiol Mol Biol Rev. 2019 Sep 4;83(4):e00034-19. doi: 10.1128/MMBR.00034-19. Print 2019 Nov 20. Microbiol Mol Biol Rev. 2019. PMID: 31484692 Free PMC article. Review.
-
The PrfA virulence regulon.Microbes Infect. 2007 Aug;9(10):1196-207. doi: 10.1016/j.micinf.2007.05.007. Epub 2007 May 7. Microbes Infect. 2007. PMID: 17764998 Review.
Cited by
-
Listeriosis: Characteristics, Occurrence in Domestic Animals, Public Health Significance, Surveillance and Control.Microorganisms. 2024 Oct 12;12(10):2055. doi: 10.3390/microorganisms12102055. Microorganisms. 2024. PMID: 39458364 Free PMC article. Review.
-
An insight into the role of branched-chain α-keto acid dehydrogenase (BKD) complex in branched-chain fatty acid biosynthesis and virulence of Listeria monocytogenes.J Bacteriol. 2024 Jul 25;206(7):e0003324. doi: 10.1128/jb.00033-24. Epub 2024 Jun 20. J Bacteriol. 2024. PMID: 38899896 Free PMC article.
-
The cAMP receptor protein from Gardnerella vaginalis is not regulated by ligands.Commun Biol. 2024 Oct 1;7(1):1233. doi: 10.1038/s42003-024-06957-1. Commun Biol. 2024. PMID: 39354127 Free PMC article.
-
Staphylococcus aureus lipid factors modulate melanoma cell clustering and invasion.Dis Model Mech. 2024 Sep 1;17(9):dmm050770. doi: 10.1242/dmm.050770. Epub 2024 Sep 16. Dis Model Mech. 2024. PMID: 39284707 Free PMC article.
-
NrnA Is a Linear Dinucleotide Phosphodiesterase with Limited Function in Cyclic Dinucleotide Metabolism in Listeria monocytogenes.J Bacteriol. 2022 Jan 18;204(1):e0020621. doi: 10.1128/JB.00206-21. Epub 2021 Oct 18. J Bacteriol. 2022. PMID: 34662239 Free PMC article.
References
-
- Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, Goebel W, Vazquez-Boland JA, European Listeria Genome Consortium. 2002. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci U S A 99:431–436. doi:10.1073/pnas.012363899. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

