Contributions of a LysR Transcriptional Regulator to Listeria monocytogenes Virulence and Identification of Its Regulons
- PMID: 32179628
- PMCID: PMC7186455
- DOI: 10.1128/JB.00087-20
Contributions of a LysR Transcriptional Regulator to Listeria monocytogenes Virulence and Identification of Its Regulons
Abstract
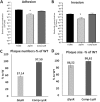
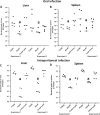
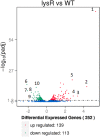
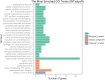
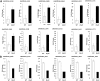
The capacity of Listeria monocytogenes to adapt to environmental changes is facilitated by a large number of regulatory proteins encoded by its genome. Among these proteins are the uncharacterized LysR-type transcriptional regulators (LTTRs). LTTRs can work as positive and/or negative transcription regulators at both local and global genetic levels. Previously, our group determined by comparative genome analysis that one member of the LTTRs (NCBI accession no. WP_003734782) was present in pathogenic strains but absent from nonpathogenic strains. The goal of the present study was to assess the importance of this transcription factor in the virulence of L. monocytogenes strain F2365 and to identify its regulons. An L. monocytogenes strain lacking lysR (the F2365ΔlysR strain) displayed significant reductions in cell invasion of and adhesion to Caco-2 cells. In plaque assays, the deletion of lysR resulted in a 42.86% decrease in plaque number and a 13.48% decrease in average plaque size. Furthermore, the deletion of lysR also attenuated the virulence of L. monocytogenes in mice following oral and intraperitoneal inoculation. The analysis of transcriptomics revealed that the transcript levels of 139 genes were upregulated, while 113 genes were downregulated in the F2365ΔlysR strain compared to levels in the wild-type bacteria. lysR-repressed genes included ABC transporters, important for starch and sucrose metabolism as well as glycerolipid metabolism, flagellar assembly, quorum sensing, and glycolysis/gluconeogenesis. Conversely, lysR activated the expression of genes related to fructose and mannose metabolism, cationic antimicrobial peptide (CAMP) resistance, and beta-lactam resistance. These data suggested that lysR contributed to L. monocytogenes virulence by broad impact on multiple pathways of gene expression.IMPORTANCEListeria monocytogenes is the causative agent of listeriosis, an infectious and fatal disease of animals and humans. In this study, we have shown that lysR contributes to Listeria pathogenesis and replication in cell lines. We also highlight the importance of lysR in regulating the transcription of genes involved in different pathways that might be essential for the growth and persistence of L. monocytogenes in the host or under nutrient limitation. Better understanding L. monocytogenes pathogenesis and the role of various virulence factors is necessary for further development of prevention and control strategies.
Keywords: Listeria; Listeria monocytogenes; RNA sequence; transcription regulator; virulence.
Copyright © 2020 American Society for Microbiology.
Figures
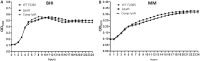

References
-
- Lampidis R, Gross R, Sokolovic Z, Goebel W, Kreft J. 1994. The virulence regulator protein of Listeria ivanovii is highly homologous to PrfA from Listeria monocytogenes and both belong to the Crp-Fnr family of transcription regulators. Mol Microbiol 13:141–151. doi:10.1111/j.1365-2958.1994.tb00409.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

