Visualizing and Manipulating Biological Processes by Using HaloTag and SNAP-Tag Technologies
- PMID: 32180315
- PMCID: PMC7367766
- DOI: 10.1002/cbic.202000037
Visualizing and Manipulating Biological Processes by Using HaloTag and SNAP-Tag Technologies
Abstract
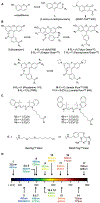
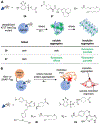
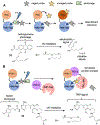
Visualizing and manipulating the behavior of proteins is crucial to understanding the physiology of the cell. Methods of biorthogonal protein labeling are important tools to attain this goal. In this review, we discuss advances in probe technology specific for self-labeling protein tags, focusing mainly on the application of HaloTag and SNAP-tag systems. We describe the latest developments in small-molecule probes that enable fluorogenic (no wash) imaging and super-resolution fluorescence microscopy. In addition, we cover several methodologies that enable the perturbation or manipulation of protein behavior and function towards the control of biological pathways. Thus, current technical advances in the HaloTag and SNAP-tag systems means that they are becoming powerful tools to enable the visualization and manipulation of biological processes, providing invaluable scientific insights that are difficult to obtain by traditional methodologies. As the multiplex of self-labeling protein tag systems continues to be developed and expanded, the utility of these protein tags will allow researchers to address previously inaccessible questions at the forefront of biology.
Keywords: HaloTag; SNAP-tag; bio-orthogonal chemistry; chemical biology; molecular probes.
© 2020 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures








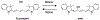
References
-
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV, ACS Chem. Biol 2008, 3, 373–382. - PubMed
-
- Gautier A, Juillerat A, Heinis C, Correa IR, Kindermann M, Beaufils F, Johnsson K, Chem. Biol 2008, 15, 128–136. - PubMed
-
- Hori Y, Ueno H, Mizukami S, Kikuchi K, J. Am. Chem. Soc 2009, 131, 16610–16611. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

