Wikidata as a knowledge graph for the life sciences
- PMID: 32180547
- PMCID: PMC7077981
- DOI: 10.7554/eLife.52614
Wikidata as a knowledge graph for the life sciences
Abstract
Wikidata is a community-maintained knowledge base that has been assembled from repositories in the fields of genomics, proteomics, genetic variants, pathways, chemical compounds, and diseases, and that adheres to the FAIR principles of findability, accessibility, interoperability and reusability. Here we describe the breadth and depth of the biomedical knowledge contained within Wikidata, and discuss the open-source tools we have built to add information to Wikidata and to synchronize it with source databases. We also demonstrate several use cases for Wikidata, including the crowdsourced curation of biomedical ontologies, phenotype-based diagnosis of disease, and drug repurposing.
Keywords: computational biology; data mining; drug repurposing; knowledge graphs; none; science forum; systems biology; wikidata.
© 2020, Waagmeester et al.
Conflict of interest statement
AW, GS, SB, BG, MG, OG, KH, HH, TH, KH, SK, MM, MM, DM, EM, AP, TP, AR, NQ, LS, TS, DS, RS, KT, GT, RT, SU, EW, CW, AS No competing interests declared
Figures
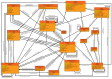
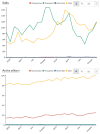

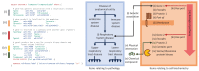


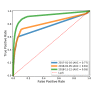
References
-
- Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bourexis D, Brister JR, Bryant SH, Canese K, Cavanaugh M, Charowhas C, Clark K, Dondoshansky I, Feolo M, Fitzpatrick L, Funk K, Geer LY, Gorelenkov V, Graeff A, Hlavina W, Holmes B, Johnson M, Kattman B, Khotomlianski V, Kimchi A, Kimelman M, Kimura M, Kitts P, Klimke W, Kotliarov A, Krasnov S, Kuznetsov A, Landrum MJ, Landsman D, Lathrop S, Lee JM, Leubsdorf C, Lu Z, Madden TL, Marchler-Bauer A, Malheiro A, Meric P, Karsch-Mizrachi I, Mnev A, Murphy T, Orris R, Ostell J, O'Sullivan C, Palanigobu V, Panchenko AR, Phan L, Pierov B, Pruitt KD, Rodarmer K, Sayers EW, Schneider V, Schoch CL, Schuler GD, Sherry ST, Siyan K, Soboleva A, Soussov V, Starchenko G, Tatusova TA, Thibaud-Nissen F, Todorov K, Trawick BW, Vakatov D, Ward M, Yaschenko E, Zasypkin A, Zbicz K, Coordinators NR, NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. - DOI - PMC - PubMed
-
- Ayers P, Mietchen D, Orlowitz J, Proffitt M, Rodlund S, Seiver E, Taraborelli D, Vershbow B. WikiCite 2018-2019: Citations for the Sum of All Human Knowledge. Wikimedia Foundation; 2019.
-
- Bastian F, Parmentier G, Roux J, Moretti S, Laudet V, Robinson-Rechavi M. Bgee: Integrating and Comparing Heterogeneous Transcriptome Data Among Species. In: Bairoch A, Cohen-Boulakia S, Froidevaux C, editors. Data Integration in the Life Sciences, Lecture Notes in Computer Science. Berlin Heidelberg: Springer; 2008. pp. 124–131. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

