Compound C Reducing Interferon Expression by Inhibiting cGAMP Accumulation
- PMID: 32180716
- PMCID: PMC7059800
- DOI: 10.3389/fphar.2020.00088
Compound C Reducing Interferon Expression by Inhibiting cGAMP Accumulation
Abstract
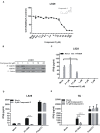
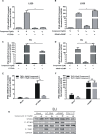
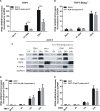
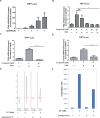
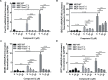
Cyclic GMP-AMP (cGAMP) synthase (cGAS) is a major DNA sensor responsible for cytosolic DNA-mediated innate immune response. Inhibition of cGAS may be an effective strategy for treating autoimmune diseases such as Aicardi-Goutieres syndrome and systemic lupus erythematosus. Compound C (also known as Dorsomorphin) has been annotated as a potent and reversible inhibitor for AMPKs as well as ALK protein kinases. Here, we report a new function of Compound C which can suppress dsDNA-dependent type I interferon induction. These effects were not dependent on the activities of AMPK proteins. In vitro assays and liquid chromatograph-mass spectrometry data show that Compound C has the capability of reducing cGAMP accumulation, suggesting that Compound C may function as a modulator involved in the cGAS-STING-mediated DNA sensing pathway. Furthermore, Compound C is able to rescue the autoimmune phenotypes in a mouse model carrying the Trex1 gene deficiency. These data demonstrate a new and inverse correlation between Compound C and type I interferon production in response to dsDNA signaling.
Keywords: DNA sensing; cGAMP; cGAS; compound C; dorsomorphin; type I interferon.
Copyright © 2020 Lai, Luo, Tian, Zhang, Huang, Wang, Li, Cai and Chen.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials

