Glypican-1 in human glioblastoma: implications in tumorigenesis and chemotherapy
- PMID: 32180897
- PMCID: PMC7061737
- DOI: 10.18632/oncotarget.27492
Glypican-1 in human glioblastoma: implications in tumorigenesis and chemotherapy
Abstract
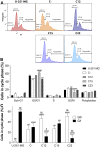
Glioblastoma is one of the most common malignant brain tumors, with which patients have a mean survival of 24 months. Glypican-1 has been previously shown to be overexpressed in human glioblastoma and to be negatively correlated with patient's survival. This study aimed to investigate how glypican-1 influences the tumoral profile of human glioblastoma using in vitro cell line models. By downregulating the expression of glypican-1 in U-251 MG cells, we observed that the cellular growth and proliferation were highly reduced, in which cells were significantly shifted towards G0 as opposed to G1 phases. Cellular migration was severely affected, and glypican-1 majorly impacted the affinity towards laminin-binding of glioblastoma U-251 MG cells. This proteoglycan was highly prevalent in glioblastoma cells, being primarily localized in the cellular membrane and extracellular vesicles, occasionally with glypican-3. Glypican-1 could also be found in cell-cell junctions with syndecan-4 but was not identified in lipid rafts in this study. Glypican-1-silenced cells were much more susceptible to temozolomide than in U-251 MG itself. Therefore, we present evidence not only to support facts that glypican-1 is an elementary macromolecule in glioblastoma tumoral microenvironment but also to introduce this proteoglycan as a promising therapeutic target for this lethal tumor.
Keywords: chemotherapy resistance; glioblastoma; glypican-1; temozolomide; tumorigenesis.
Conflict of interest statement
CONFLICTS OF INTEREST The authors report no competing interests.
Figures







References
-
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016; 131:803–20. 10.1007/s00401-016-1545-1. - DOI - PubMed
LinkOut - more resources
Full Text Sources

