Polycysteine-encoding leaderless short ORFs function as cysteine-responsive attenuators of operonic gene expression in mycobacteria
- PMID: 32181921
- PMCID: PMC8764745
- DOI: 10.1111/mmi.14498
Polycysteine-encoding leaderless short ORFs function as cysteine-responsive attenuators of operonic gene expression in mycobacteria
Abstract
Genome-wide transcriptomic analyses have revealed abundant expressed short open reading frames (ORFs) in bacteria. Whether these short ORFs, or the small proteins they encode, are functional remains an open question. One quarter of mycobacterial mRNAs are leaderless, beginning with a 5'-AUG or GUG initiation codon. Leaderless mRNAs often encode unannotated short ORFs as the first gene of a polycistronic transcript. Here, we show that polycysteine-encoding leaderless short ORFs function as cysteine-responsive attenuators of operonic gene expression. Detailed mutational analysis shows that one polycysteine short ORF controls expression of the downstream genes. Our data indicate that ribosomes stalled in the polycysteine tract block mRNA structures that otherwise sequester the ribosome-binding site of the 3'gene. We assessed endogenous proteomic responses to cysteine limitation in Mycobacterium smegmatis using mass spectrometry. Six cysteine metabolic loci having unannotated polycysteine-encoding leaderless short ORF architectures responded to cysteine limitation, revealing widespread cysteine-responsive attenuation in mycobacteria. Individual leaderless short ORFs confer independent operon-level control, while their shared dependence on cysteine ensures a collective response mediated by ribosome pausing. We propose the term ribulon to classify ribosome-directed regulons. Regulon-level coordination by ribosomes on sensory short ORFs illustrates one utility of the many unannotated short ORFs expressed in bacterial genomes.
Keywords: cysteine; mycobacteria; operon; polycysteine; regulon; short ORFs.
© 2020 John Wiley & Sons Ltd.
Figures

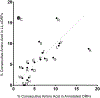
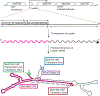






Similar articles
-
Pervasive translation in Mycobacterium tuberculosis.Elife. 2022 Mar 28;11:e73980. doi: 10.7554/eLife.73980. Elife. 2022. PMID: 35343439 Free PMC article.
-
The Impact of Leadered and Leaderless Gene Structures on Translation Efficiency, Transcript Stability, and Predicted Transcription Rates in Mycobacterium smegmatis.J Bacteriol. 2020 Apr 9;202(9):e00746-19. doi: 10.1128/JB.00746-19. Print 2020 Apr 9. J Bacteriol. 2020. PMID: 32094162 Free PMC article.
-
Adaptation of mycobacteria to growth conditions: a theoretical analysis of changes in gene expression revealed by microarrays.PLoS One. 2013 Apr 12;8(4):e59883. doi: 10.1371/journal.pone.0059883. Print 2013. PLoS One. 2013. PMID: 23593152 Free PMC article.
-
The Emerging World of Small ORFs.Trends Plant Sci. 2016 Apr;21(4):317-328. doi: 10.1016/j.tplants.2015.11.005. Epub 2015 Dec 10. Trends Plant Sci. 2016. PMID: 26684391 Review.
-
Translation initiation at AUG and non-AUG triplets in plants.Plant Sci. 2023 Oct;335:111822. doi: 10.1016/j.plantsci.2023.111822. Epub 2023 Aug 14. Plant Sci. 2023. PMID: 37574140 Review.
Cited by
-
In vitro studies of the protein-interaction network of cell-wall lytic transglycosylase RlpA of Pseudomonas aeruginosa.Commun Biol. 2022 Nov 30;5(1):1314. doi: 10.1038/s42003-022-04230-x. Commun Biol. 2022. PMID: 36451021 Free PMC article.
-
The BR-body proteome contains a complex network of protein-protein and protein-RNA interactions.Cell Rep. 2023 Oct 31;42(10):113229. doi: 10.1016/j.celrep.2023.113229. Epub 2023 Oct 19. Cell Rep. 2023. PMID: 37815915 Free PMC article.
-
Pervasive translation in Mycobacterium tuberculosis.Elife. 2022 Mar 28;11:e73980. doi: 10.7554/eLife.73980. Elife. 2022. PMID: 35343439 Free PMC article.
-
Mycobacterium smegmatis: The Vanguard of Mycobacterial Research.J Bacteriol. 2023 Jan 26;205(1):e0033722. doi: 10.1128/jb.00337-22. Epub 2023 Jan 4. J Bacteriol. 2023. PMID: 36598232 Free PMC article. Review.
-
Proteo-genetic analysis reveals clear hierarchy of ESX-1 secretion in Mycobacterium marinum.Proc Natl Acad Sci U S A. 2022 Jun 14;119(24):e2123100119. doi: 10.1073/pnas.2123100119. Epub 2022 Jun 7. Proc Natl Acad Sci U S A. 2022. PMID: 35671426 Free PMC article.
References
-
- BECHHOFER DH 1990. Triple post-transcriptional control. Mol Microbiol, 4, 1419–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

