Loop-closure kinetics reveal a stable, right-handed DNA intermediate in Cre recombination
- PMID: 32182357
- PMCID: PMC7192630
- DOI: 10.1093/nar/gkaa153
Loop-closure kinetics reveal a stable, right-handed DNA intermediate in Cre recombination
Abstract
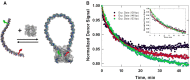
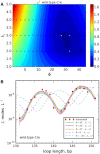
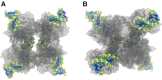
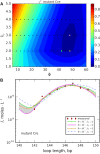
In Cre site-specific recombination, the synaptic intermediate is a recombinase homotetramer containing a pair of loxP DNA target sites. The enzyme system's strand-exchange mechanism proceeds via a Holliday-junction (HJ) intermediate; however, the geometry of DNA segments in the synapse has remained highly controversial. In particular, all crystallographic structures are consistent with an achiral, planar Holliday-junction (HJ) structure, whereas topological assays based on Cre-mediated knotting of plasmid DNAs are consistent with a right-handed chiral junction. We use the kinetics of loop closure involving closely spaced (131-151 bp) loxP sites to investigate the in-aqueo ensemble of conformations for the longest-lived looped DNA intermediate. Fitting the experimental site-spacing dependence of the loop-closure probability, J, to a statistical-mechanical theory of DNA looping provides evidence for substantial out-of-plane HJ distortion, which unequivocally stands in contrast to the square-planar intermediate geometry from Cre-loxP crystal structures and those of other int-superfamily recombinases. J measurements for an HJ-isomerization-deficient Cre mutant suggest that the apparent geometry of the wild-type complex is consistent with temporal averaging of right-handed and achiral structures. Our approach connects the static pictures provided by crystal structures and the natural dynamics of macromolecules in solution, thus advancing a more comprehensive dynamic analysis of large nucleoprotein structures and their mechanisms.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
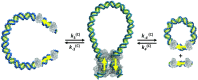

References
-
- Castán A., Hernández P., Krimer D.B., Schvartzman J.B.. Drolet M. DNA catenation reveals the dynamics of DNA topology during replication. DNA Topoisomerases: Methods and Protocols. 2018; NY: Springer New York; 75–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

