G-quadruplex Stabilization Fuels the ALT Pathway in ALT-positive Osteosarcoma Cells
- PMID: 32183119
- PMCID: PMC7140816
- DOI: 10.3390/genes11030304
G-quadruplex Stabilization Fuels the ALT Pathway in ALT-positive Osteosarcoma Cells
Abstract
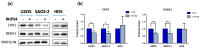
Most human tumors maintain telomere lengths by telomerase, whereas a portion of them (10%-15%) uses a mechanism named alternative lengthening of telomeres (ALT). The telomeric G-quadruplex (G4) ligand RHPS4 is known for its potent antiproliferative effect, as shown in telomerase-positive cancer models. Moreover, RHPS4 is also able to reduce cell proliferation in ALT cells, although the influence of G4 stabilization on the ALT mechanism has so far been poorly investigated. Here we show that sensitivity to RHPS4 is comparable in ALT-positive (U2OS; SAOS-2) and telomerase-positive (HOS) osteosarcoma cell lines, unlinking the telomere maintenance mechanism and RHPS4 responsiveness. To investigate the impact of G4 stabilization on ALT, the cardinal ALT hallmarks were analyzed. A significant induction of telomeric doublets, telomeric clusterized DNA damage, ALT-associated Promyelocytic Leukaemia-bodies (APBs), telomere sister chromatid exchanges (T-SCE) and c-circles was found exclusively in RHPS4-treated ALT cells. We surmise that RHPS4 affects ALT mechanisms through the induction of replicative stress that in turn is converted in DNA damage at telomeres, fueling recombination. In conclusion, our work indicates that RHPS4-induced telomeric DNA damage promotes overactivation of telomeric recombination in ALT cells, opening new questions on the therapeutic employment of G4 ligands in the treatment of ALT positive tumors.
Keywords: ALT; G-quadruplex; RHPS4; replicative stress; telomeres.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Blackburn E.H., Chan S., Chang J., Fulton T.B., Krauskopf A., McEachern M., Prescott J., Roy J., Smith C., Wang H. Cold Spring Harbor Symposia on Quantitative Biology. Volume 65. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2000. Molecular manifestations and molecular determinants of telomere capping; pp. 253–264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

