Lactic Acid Bacteria: Variability Due to Different Pork Breeds, Breeding Systems and Fermented Sausage Production Technology
- PMID: 32183247
- PMCID: PMC7142627
- DOI: 10.3390/foods9030338
Lactic Acid Bacteria: Variability Due to Different Pork Breeds, Breeding Systems and Fermented Sausage Production Technology
Abstract
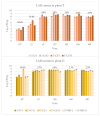
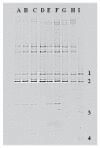
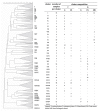
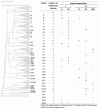
Changes in the ecology of the various lactic acid bacteria (LAB) species, which are involved in traditional fermented sausages, were investigated in the light of the use of different breeds of pork, each of which was raised in two different environments and processed using two different technologies. The semi-quantitative molecular method was applied in order to understand how the different species alternate over time, as well as their concentration ratios. A significant increase in LAB over the first days of fermentation characterized the trials where the starter culture wasn't added (T), reaching values of 107-108 cfu g-1. On the other hand, in the trials in which sausages were produced with starter addition, LAB counts had a less significant incremental jump from about 106 cfu g-1 (concentration of the inoculum) to 108 cfu g-1. Lactobacillus sakei and Lb. curvatus were detected as the prevalent population in all the observed fermentations. Pediococcus pentosaceus, Lb. casei, Leuconostoc mesenteroides, Lactococcus garviae, and Lb. graminis also appeared, but their concentration ratios varied depending on the diverse experimental settings. The results of cluster analysis showed that a plant- and breed-specific LAB ecology exists. In addition, it was also observed that the breeding system can influence the presence of certain LAB species.
Keywords: breed; breeding system; ecology; fermented sausages; lactic acid bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Cocolin L., Manzano M., Cantoni C., Comi G. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italia sausages. Appl. Environ. Microbiol. 2001;67:5113–6282. doi: 10.1128/AEM.67.11.5113-5121.2001. - DOI - PMC - PubMed
-
- Lücke F.K. Fermented sausages. In: Wood B.J.B., editor. Microbiology of Fermented Food. Elsevier Applied Science; New York, NY, USA: 1985.
-
- Iacumin L., Osualdini M., Bovolenta S., Boscolo D., Chiesa L., Panseri S., Comi G. Microbial, chemico-physical and volatile aromatic compounds characterization of Pitina PGI, a peculiar sausage-like product of North East Italy. Meat Sci. 2020;162:108081. doi: 10.1016/j.meatsci.2020.108081. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

