GsMAS1 Encoding a MADS-box Transcription Factor Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana
- PMID: 32183485
- PMCID: PMC7139582
- DOI: 10.3390/ijms21062004
GsMAS1 Encoding a MADS-box Transcription Factor Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana
Abstract
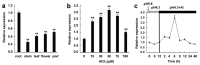
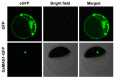
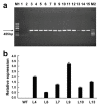
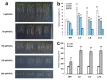
The MADS-box transcription factors (TFs) are essential in regulating plant growth and development, and conferring abiotic and metal stress resistance. This study aims to investigate GsMAS1 function in conferring tolerance to aluminum stress in Arabidopsis. The GsMAS1 from the wild soybean BW69 line encodes a MADS-box transcription factor in Glycine soja by bioinformatics analysis. The putative GsMAS1 protein was localized in the nucleus. The GsMAS1 gene was rich in soybean roots presenting a constitutive expression pattern and induced by aluminum stress with a concentration-time specific pattern. The analysis of phenotypic observation demonstrated that overexpression of GsMAS1 enhanced the tolerance of Arabidopsis plants to aluminum (Al) stress with larger values of relative root length and higher proline accumulation compared to those of wild type at the AlCl3 treatments. The genes and/or pathways regulated by GsMAS1 were further investigated under Al stress by qRT-PCR. The results indicated that six genes resistant to Al stress were upregulated, whereas AtALMT1 and STOP2 were significantly activated by Al stress and GsMAS1 overexpression. After treatment of 50 μM AlCl3, the RNA abundance of AtALMT1 and STOP2 went up to 17-fold and 37-fold than those in wild type, respectively. Whereas the RNA transcripts of AtALMT1 and STOP2 were much higher than those in wild type with over 82% and 67% of relative expression in GsMAS1 transgenic plants, respectively. In short, the results suggest that GsMAS1 may increase resistance to Al toxicity through certain pathways related to Al stress in Arabidopsis.
Keywords: Al stress; Arabidopsis thaliana; Glycine Soja; GsMAS1; MADS.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Heterologous Expression of a Glycine soja C2H2 Zinc Finger Gene Improves Aluminum Tolerance in Arabidopsis.Int J Mol Sci. 2020 Apr 15;21(8):2754. doi: 10.3390/ijms21082754. Int J Mol Sci. 2020. PMID: 32326652 Free PMC article.
-
GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in Arabidopsis thaliana.BMC Plant Biol. 2018 Sep 29;18(1):212. doi: 10.1186/s12870-018-1397-z. BMC Plant Biol. 2018. PMID: 30268093 Free PMC article.
-
GsERF1 enhances Arabidopsis thaliana aluminum tolerance through an ethylene-mediated pathway.BMC Plant Biol. 2022 May 24;22(1):258. doi: 10.1186/s12870-022-03625-6. BMC Plant Biol. 2022. PMID: 35610574 Free PMC article.
-
Transcriptional Regulation of Aluminum-Tolerance Genes in Higher Plants: Clarifying the Underlying Molecular Mechanisms.Front Plant Sci. 2017 Aug 8;8:1358. doi: 10.3389/fpls.2017.01358. eCollection 2017. Front Plant Sci. 2017. PMID: 28848571 Free PMC article. Review.
-
Root Adaptation via Common Genetic Factors Conditioning Tolerance to Multiple Stresses for Crops Cultivated on Acidic Tropical Soils.Front Plant Sci. 2020 Nov 12;11:565339. doi: 10.3389/fpls.2020.565339. eCollection 2020. Front Plant Sci. 2020. PMID: 33281841 Free PMC article. Review.
Cited by
-
Aluminum Stress Response Is Regulated Through a miR156/SPL13 Module in Medicago sativa.Genes (Basel). 2025 Jun 27;16(7):751. doi: 10.3390/genes16070751. Genes (Basel). 2025. PMID: 40725407 Free PMC article.
-
Two Sweet Sorghum (Sorghum bicolor L.) WRKY Transcription Factors Promote Aluminum Tolerance via the Reduction in Callose Deposition.Int J Mol Sci. 2023 Jun 17;24(12):10288. doi: 10.3390/ijms241210288. Int J Mol Sci. 2023. PMID: 37373435 Free PMC article.
-
ZmNRAMP4 Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana.Int J Mol Sci. 2022 Jul 25;23(15):8162. doi: 10.3390/ijms23158162. Int J Mol Sci. 2022. PMID: 35897738 Free PMC article.
-
GsMYB7 encoding a R2R3-type MYB transcription factor enhances the tolerance to aluminum stress in soybean (Glycine max L.).BMC Genomics. 2022 Jul 22;23(1):529. doi: 10.1186/s12864-022-08744-w. BMC Genomics. 2022. PMID: 35869448 Free PMC article.
-
Functional Characterization of Aluminum (Al)-Responsive Membrane-Bound NAC Transcription Factors in Soybean Roots.Int J Mol Sci. 2021 Nov 27;22(23):12854. doi: 10.3390/ijms222312854. Int J Mol Sci. 2021. PMID: 34884659 Free PMC article.
References
-
- Hoekenga O.A., Vision T.J., Shaff J.E., Monforte A.J., Lee G.P., Howell S.H., Kochian L.V. Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsbergerecta × Columbia) by quantitative trait locus mapping. A physiologically simple but genetically complex trait. Plant Physiol. 2003;132:936–948. doi: 10.1104/pp.103.023085. - DOI - PMC - PubMed
-
- Pandey P., Srivastava R.K., Dubey R.S. Salicylic acid alleviates aluminum toxicity in rice seedlings better than magnesium and calcium by reducing aluminum uptake, suppressing oxidative damage and increasing antioxidative defense. Ecotoxicology. 2013;22:656–670. doi: 10.1007/s10646-013-1058-9. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- the Major Project of New Varieties Cultivation of Genetically Modified Organisms (2016ZX08004002-007), the National Natural Science Foundation of China (31771816, 31971965), the Key Projects of International Scientific and Technological Innovation Coopera/Qibin MA
- the China Agricultural Research System (CARS-04-PS09), the Project of Science and Technology of Guangzhou (201804020015), the Research Project of the State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (4100-M13024)/Hai Nian
LinkOut - more resources
Full Text Sources

