Screening a chemically defined extracellular matrix mimetic substrate library to identify substrates that enhance substrate-mediated transfection
- PMID: 32183552
- PMCID: PMC7153211
- DOI: 10.1177/1535370220913501
Screening a chemically defined extracellular matrix mimetic substrate library to identify substrates that enhance substrate-mediated transfection
Abstract
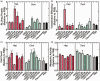
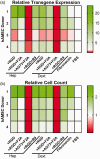
Nonviral gene delivery, though limited by inefficiency, has extensive utility in cell therapy, tissue engineering, and diagnostics. Substrate-mediated gene delivery (SMD) increases efficiency and allows transfection at a cell-biomaterial interface, by immobilizing and concentrating nucleic acid complexes on a surface. Efficient SMD generally requires substrates to be coated with serum or other protein coatings to mediate nucleic acid complex immobilization, as well as cell adhesion and growth; however, this strategy limits reproducibility and may be difficult to translate for clinical applications. As an alternative, we screened a chemically defined combinatorial library of 20 different extracellular matrix mimetic substrates containing combinations of (1) different sulfated polysaccharides that are essential extracellular matrix glycosaminoglycans (GAGs), with (2) mimetic peptides derived from adhesion proteins, growth factors, and cell-penetrating domains, for use as SMD coatings. We identified optimal substrates for DNA lipoplex and polyplex SMD transfection of fibroblasts and human mesenchymal stem cells. Optimal extracellular matrix mimetic substrates varied between cell type, donor source, and transfection reagent, but typically contained Heparin GAG and an adhesion peptide. Multiple substrates significantly increased transgene expression (i.e. 2- to 20-fold) over standard protein coatings. Considering previous research of similar ligands, we hypothesize extracellular matrix mimetic substrates modulate cell adhesion, proliferation, and survival, as well as plasmid internalization and trafficking. Our results demonstrate the utility of screening combinatorial extracellular matrix mimetic substrates for optimal SMD transfection towards application- and patient-specific technologies.
Impact statement: Substrate-mediated gene delivery (SMD) approaches have potential for modification of cells in applications where a cell-material interface exists. Conventional SMD uses ill-defined serum or protein coatings to facilitate immobilization of nucleic acid complexes, cell attachment, and subsequent transfection, which limits reproducibility and clinical utility. As an alternative, we screened a defined library of extracellular matrix mimetic substrates containing combinations of different glycosaminoglycans and bioactive peptides to identify optimal substrates for SMD transfection of fibroblasts and human mesenchymal stem cells. This strategy could be utilized to develop substrates for specific SMD applications in which variability exists between different cell types and patient samples.
Keywords: Mimetic peptides; extracellular matrix; glycosaminoglycans; human mesenchymal stem cells; substrate-mediated gene delivery; transfection.
Figures



References
-
- Hagstrom J. Self-assembling complexes for in vivo gene delivery. Curr Opin Mol Ther 2000; 2:143–9 - PubMed
-
- Pannier AK, Kozisek T, Segura T. Surface-and hydrogel-mediated delivery of nucleic acid nanoparticles. Nanotechnology for nucleic acid delivery. Berlin, Germany: Springer, 2019, pp.177–97 - PubMed
-
- Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug Chem 2002; 13:621–9 - PubMed
-
- Zheng J, Manuel WS, Hornsby PJ. Transfection of cells mediated by biodegradable polymer materials with surface‐bound polyethyleneimine. Biotechnol Prog 2000; 16:254–7 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

