m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer
- PMID: 32183948
- PMCID: PMC7141420
- DOI: 10.1016/j.ccell.2020.02.004
m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer
Abstract
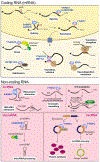
N6-Methyladenosine (m6A) RNA modification has emerged in recent years as a new layer of regulatory mechanism controlling gene expression in eukaryotes. As a reversible epigenetic modification found not only in messenger RNAs but also in non-coding RNAs, m6A affects the fate of the modified RNA molecules and plays important roles in almost all vital bioprocesses, including cancer development. Here we review the up-to-date knowledge of the pathological roles and underlying molecular mechanism of m6A modifications (in both coding and non-coding RNAs) in cancer pathogenesis and drug response/resistance, and discuss the therapeutic potential of targeting m6A regulators for cancer therapy.
Keywords: N(6)-methyladenosine (m(6)A); RNA modification; cancer epigenetics; cancer stem cells; drug resistance; epitranscriptome; immune therapy; non-coding RNA; prognosis; targeted therapeutics.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests J.C. is the scientific founder of Genovel Biotech Corp. and holds equities with the company.
Figures


References
-
- Adams JM, and Cory S (1975). Modified nucleosides and bizarre 5'-termini in mouse myeloma mRNA. Nature 255, 28–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

