An Upstream Open Reading Frame Represses Translation of Chicken PPARγ Transcript Variant 1
- PMID: 32184808
- PMCID: PMC7058706
- DOI: 10.3389/fgene.2020.00165
An Upstream Open Reading Frame Represses Translation of Chicken PPARγ Transcript Variant 1
Abstract
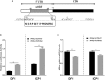
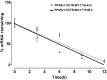
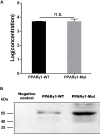
Peroxisome proliferator-activated receptor γ (PPARγ) is a master regulator of adipogenesis. The PPARγ gene produces various transcripts with different 5'-untranslated regions (5' UTRs) because of alternative promoter usage and splicing. The 5' UTR plays important roles in posttranscriptional gene regulation. However, to date, the regulatory role and underlying mechanism of 5' UTRs in the posttranscriptional regulation of PPARγ expression remain largely unclear. In this study, we investigated the effects of 5' UTRs on posttranscriptional regulation using reporter assays. Our results showed that the five PPARγ 5' UTRs exerted different effects on reporter gene activity. Bioinformatics analysis showed that chicken PPARγ transcript 1 (PPARγ1) possessed an upstream open reading frame (uORF) in its 5' UTR. Mutation analysis showed that a mutation in the uORF led to increased Renilla luciferase activity and PPARγ protein expression, but decreased Renilla luciferase and PPARγ1 mRNA expression. mRNA stability analysis using real-time RT-PCR showed that the uORF mutation did not interfere with mRNA stability, but promoter activity analysis of the cloned 5' UTR showed that the uORF mutation reduced promoter activity. Furthermore, in vitro transcription/translation assays demonstrated that the uORF mutation markedly increased the translation of PPARγ1 mRNA. Collectively, our results indicate that the uORF represses the translation of chicken PPARγ1 mRNA.
Keywords: 5′-untranslated region; PPARγ; gene expression; translational repression; upstream open reading frame.
Copyright © 2020 Chu, Huang, Ma, Cui, Yan, Li and Wang.
Figures
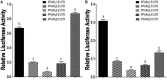




References
LinkOut - more resources
Full Text Sources

