Discrete analysis of camelid variable domains: sequences, structures, and in-silico structure prediction
- PMID: 32185102
- PMCID: PMC7061911
- DOI: 10.7717/peerj.8408
Discrete analysis of camelid variable domains: sequences, structures, and in-silico structure prediction
Abstract
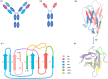
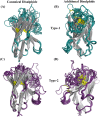
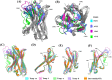
Antigen binding by antibodies requires precise orientation of the complementarity- determining region (CDR) loops in the variable domain to establish the correct contact surface. Members of the family Camelidae have a modified form of immunoglobulin gamma (IgG) with only heavy chains, called Heavy Chain only Antibodies (HCAb). Antigen binding in HCAbs is mediated by only three CDR loops from the single variable domain (VHH) at the N-terminus of each heavy chain. This feature of the VHH, along with their other important features, e.g., easy expression, small size, thermo-stability and hydrophilicity, made them promising candidates for therapeutics and diagnostics. Thus, to design better VHH domains, it is important to thoroughly understand their sequence and structure characteristics and relationship. In this study, sequence characteristics of VHH domains have been analysed in depth, along with their structural features using innovative approaches, namely a structural alphabet. An elaborate summary of various studies proposing structural models of VHH domains showed diversity in the algorithms used. Finally, a case study to elucidate the differences in structural models from single and multiple templates is presented. In this case study, along with the above-mentioned aspects of VHH, an exciting view of various factors in structure prediction of VHH, like template framework selection, is also discussed.
Keywords: Antibodies; Complementarity determining regions; Frameworks; Nanobodies; Secondary structure; Sequence structure relationship; Structural alphabet.
©2020 Melarkode Vattekatte et al.
Conflict of interest statement
Frederic Cadet is associated with PEACCEL, Paris, France. Jean-Christophe Gelly and Alexandre G. de Brevern are associated with IBL, Paris, France. Jean-Philippe Meyneil is employed by ISoft, Paris, France. Alain Malpertuy is employed by Atragene, Paris, France. Nicolas K. Shinada is sponsored by Discngine, Paris, France and ANRT, France. All other authors declare no competing interests.
Figures




References
-
- Ablynx Ablynx 2016 Annual report 2016
LinkOut - more resources
Full Text Sources
Other Literature Sources

