Genome-wide investigation of WRKY transcription factors in Tartary buckwheat (Fagopyrum tataricum) and their potential roles in regulating growth and development
- PMID: 32185114
- PMCID: PMC7060923
- DOI: 10.7717/peerj.8727
Genome-wide investigation of WRKY transcription factors in Tartary buckwheat (Fagopyrum tataricum) and their potential roles in regulating growth and development
Abstract
Background: The WRKY gene family plays important roles in plant biological functions and has been identified in many plant species. With the publication of the Tartary buckwheat genome, the evolutionary characteristics of the WRKY gene family can be systematically explored and the functions of Fagopyrum tataricum WRKY (FtWRKY) genes in the growth and development of this plant also can be predicted.
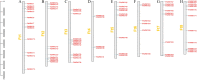
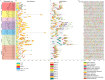
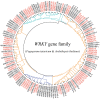
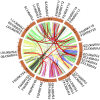
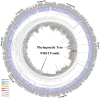
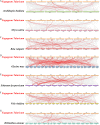
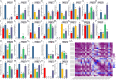
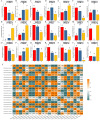
Methods: In this study, the FtWRKY genes were identified by the BLASTP method, and HMMER, SMART, Pfam and InterPro were used to determine whether the FtWRKY genes contained conserved domains. The phylogenetic trees including FtWRKY and WRKY genes in other plants were constructed by the neighbor-joining (NJ) and maximum likelihood (ML) methods. The intron and exon structures of the FtWRKY genes were analyzed by the gene structure display server, and the motif compositions were analyzed by MEME. Chromosome location information of FtWRKY genes was obtained with gff files and sequencing files, and visualized by Circos, and the collinear relationship was analyzed by Dual synteny plotter software. The expression levels of 26 FtWRKY genes from different groups in roots, leaves, flowers, stems and fruits at the green fruit, discoloration and initial maturity stage were measured through quantitative real-time polymerase chain reaction (qRT-PCR) analysis.
Results: A total of 76 FtWRKY genes identified from the Tartary buckwheat genome were divided into three groups. FtWRKY genes in the same group had similar gene structures and motif compositions. Despite the lack of tandem-duplicated gene pairs, there were 23 pairs of segmental-duplicated gene pairs. The synteny gene pairs of FtWRKY genes and Glycine max WRKY genes were the most. FtWRKY42 was highly expressed in roots and may perform similar functions as its homologous gene AtWRKY75, playing a role in lateral root and hairy root formation. FtWRKY9, FtWRKY42 and FtWRKY60 were highly expressed in fruits and may play an important role in fruit development.
Conclusion: We have identified several candidate FtWRKY genes that may perform critical functions in the development of Tartary buckwheat root and fruit, which need be verified through further research. Our study provides useful information on WRKY genes in regulating growth and development and establishes a foundation for screening WRKY genes to improve Tartary buckwheat quality.
Keywords: Development; Fruit; Tartary buckwheat genome; FtWRKY.
©2020 Sun et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
References
-
- Bonafaccia G, Marocchini M, Kreft I. Composition and technological properties of the flour and bran from common and tartary buckwheat. Food Chemistry. 2003;80:9–15. doi: 10.1016/S0308-8146(02)00228-5. - DOI
LinkOut - more resources
Full Text Sources

