Discovery of non-HLA antibodies associated with cardiac allograft rejection and development and validation of a non-HLA antigen multiplex panel: From bench to bedside
- PMID: 32185871
- PMCID: PMC7494540
- DOI: 10.1111/ajt.15863
Discovery of non-HLA antibodies associated with cardiac allograft rejection and development and validation of a non-HLA antigen multiplex panel: From bench to bedside
Abstract
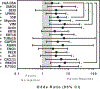
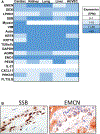
We analyzed humoral immune responses to nonhuman leukocyte antigen (HLA) after cardiac transplantation to identify antibodies associated with allograft rejection. Protein microarray identified 366 non-HLA antibodies (>1.5 fold, P < .5) from a discovery cohort of HLA antibody-negative, endothelial cell crossmatch-positive sera obtained from 12 cardiac allograft recipients at the time of biopsy-proven rejection. From these, 19 plasma membrane proteins and 10 autoantigens identified from gene ontology analysis were combined with 48 proteins identified through literature search to generate a multiplex bead array. Longitudinal sera from a multicenter cohort of adult cardiac allograft recipients (samples: n = 477 no rejection; n = 69 rejection) identified 18 non-HLA antibodies associated with rejection (P < .1) including 4 newly identified non-HLA antigenic targets (DEXI, EMCN, LPHN1, and SSB). CART analysis showed 5/18 non-HLA antibodies distinguished rejection vs nonrejection. Antibodies to 4/18 non-HLA antigens synergize with HLA donor-specific antibodies and significantly increase the odds of rejection (P < .1). The non-HLA panel was validated using an independent adult cardiac transplant cohort (n = 21 no rejection; n = 42 rejection, >1R) with an area under the curve of 0.87 (P < .05) with 92.86% sensitivity and 66.67% specificity. We conclude that multiplex bead array assessment of non-HLA antibodies identifies cardiac transplant recipients at risk of rejection.
Keywords: autoantibody; autoantigen; clinical research/practice; heart transplantation/cardiology; histocompatibility; immunogenetics; microarray/protein array; organ transplantation in general; rejection; translational research/science.
© 2020 The American Society of Transplantation and the American Society of Transplant Surgeons.
Conflict of interest statement
DISCLOSURE
The authors of this manuscript have conflicts of interest to disclose as described by the
Figures

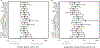




Comment in
-
Rejection in the setting of non-HLA antibody: New tools for navigating bench to bedside.Am J Transplant. 2020 Oct;20(10):2639-2641. doi: 10.1111/ajt.15975. Epub 2020 May 25. Am J Transplant. 2020. PMID: 32372531 No abstract available.
References
-
- Stehlik J, Edwards LB, Kucheryavaya AY, et al. The Registry of the International Society for Heart and Lung Transplantation: 29th official adult heart transplant report–2012. J Heart Lung Transplant. 2012;31(10):1052–1064. - PubMed
-
- Colvin-Adams M, Smith JM, Heubner BM, et al. OPTN/SRTR 2013 annual data report: heart. Am J Transplant. 2015;15(Suppl 2):1–28. - PubMed
-
- Tible M, Loupy A, Vernerey D, et al. Pathologic classification of antibody-mediated rejection correlates with donor-specific antibodies and endothelial cell activation. J Heart Lung Transplant. 2013;32(8):769–776. - PubMed
-
- Tait BD, Süsal C, Gebel HM, et al. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation. 2013;95(1):19–47. - PubMed

