Long non‑coding RNA EBLN3P promotes the recovery of the function of impaired spiral ganglion neurons by competitively binding to miR‑204‑5p and regulating TMPRSS3 expression
- PMID: 32186779
- PMCID: PMC7169660
- DOI: 10.3892/ijmm.2020.4545
Long non‑coding RNA EBLN3P promotes the recovery of the function of impaired spiral ganglion neurons by competitively binding to miR‑204‑5p and regulating TMPRSS3 expression
Abstract
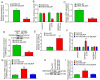
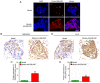
Sensorineural hearing loss (SNHL) is one of the major leading causes of hearing impairment, and is typically characterized by the degeneration of spiral ganglion neurons (SGNs). In previous studies by the authors, it was demonstrated that microRNA (miRNA or miR)‑204‑5p decreased the viability of SGNs by inhibiting the expression of transmembrane protease, serine 3 (TMPRSS3), which was closely associated with the development of SGNs. However, the upstream regulatory mechanism of miR‑204‑5p was not fully elucidated. The present study found that an important upstream regulatory factor of miR‑204‑5p, long non‑coding RNA (lncRNA) EBLN3P, was expressed at low levels in impaired SGNs, whereas it was expressed at high levels in normal SGNs. Mechanistic analyses demonstrated that lncRNA EBLN3P functioned as a competing endogenous RNA (ceRNA) when regulating miR‑204‑5p in normal SGNs. In addition, lncRNA EBLN3P regulated TMPRSS3 expression via the regulation of miR‑204‑5p in normal SGNs. In vitro functional analysis revealed that lncRNA EBLN3P promoted the recovery of the viability of normal SGNs and inhibited the apoptosis of normal SGNs. Finally, the results revealed a recovery‑promoting effect of lncRNA EBLN3P on the structure and function of impaired SGNs in models of deafness. On the whole, the findings of the present study demonstrate that lncRNA EBLN3P promotes the recovery of the function of impaired SGNs by competitively binding to miR‑204‑5p and regulating TMPRSS3 expression. This suggests that lncRNA EBLN3P may be a potential therapeutic target for diseases involving SNHL.
Figures




References
-
- Charizopoulou N, Lelli A, Schraders M, Ray K, Hildebrand MS, Ramesh A, Srisailapathy CR, Oostrik J, Admiraal RJ, Neely HR, et al. Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat Commun. 2011;2:201. doi: 10.1038/ncomms1200. - DOI - PMC - PubMed
-
- Clark GM. Cochlear Implants: Fundamentals and applications Graeme Clark. Springer; New York: 2003. - DOI
-
- Feghali JG, Lefebvre PP, Staecker H, Kopke R, Frenz DA, Malgrange B, Liu W, Moonen G, Ruben RJ, Van de Water TR. Mammalian auditory hair cell regeneration/repair and protection: A review and future directions. Ear Nose. Throat J. 1998;77:276–280. 282–285. doi: 10.1177/014556139807700409. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

