Inhibition of transcription leads to rewiring of locus-specific chromatin proteomes
- PMID: 32188699
- PMCID: PMC7197482
- DOI: 10.1101/gr.256255.119
Inhibition of transcription leads to rewiring of locus-specific chromatin proteomes
Abstract
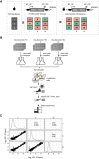
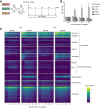
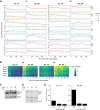
Transcription of a chromatin template involves the concerted interaction of many different proteins and protein complexes. Analyses of specific factors showed that these interactions change during stress and upon developmental switches. However, how the binding of multiple factors at any given locus is coordinated has been technically challenging to investigate. Here we used Epi-Decoder in yeast to systematically decode, at one transcribed locus, the chromatin binding changes of hundreds of proteins in parallel upon perturbation of transcription. By taking advantage of improved Epi-Decoder libraries, we observed broad rewiring of local chromatin proteomes following chemical inhibition of RNA polymerase. Rapid reduction of RNA polymerase II binding was accompanied by reduced binding of many other core transcription proteins and gain of chromatin remodelers. In quiescent cells, where strong transcriptional repression is induced by physiological signals, eviction of the core transcriptional machinery was accompanied by the appearance of quiescent cell-specific repressors and rewiring of the interactions of protein-folding factors and metabolic enzymes. These results show that Epi-Decoder provides a powerful strategy for capturing the temporal binding dynamics of multiple chromatin proteins under varying conditions and cell states. The systematic and comprehensive delineation of dynamic local chromatin proteomes will greatly aid in uncovering protein-protein relationships and protein functions at the chromatin template.
© 2020 Poramba-Liyanage et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
