Gasdermin E suppresses tumour growth by activating anti-tumour immunity
- PMID: 32188940
- PMCID: PMC7123794
- DOI: 10.1038/s41586-020-2071-9
Gasdermin E suppresses tumour growth by activating anti-tumour immunity
Abstract
Cleavage of the gasdermin proteins to produce pore-forming amino-terminal fragments causes inflammatory cell death (pyroptosis)1. Gasdermin E (GSDME, also known as DFNA5)-mutated in familial ageing-related hearing loss2-can be cleaved by caspase 3, thereby converting noninflammatory apoptosis to pyroptosis in GSDME-expressing cells3-5. GSDME expression is suppressed in many cancers, and reduced GSDME levels are associated with decreased survival as a result of breast cancer2,6, suggesting that GSDME might be a tumour suppressor. Here we show that 20 of 22 tested cancer-associated GSDME mutations reduce GSDME function. In mice, knocking out Gsdme in GSDME-expressing tumours enhances, whereas ectopic expression in Gsdme-repressed tumours inhibits, tumour growth. This tumour suppression is mediated by killer cytotoxic lymphocytes: it is abrogated in perforin-deficient mice or mice depleted of killer lymphocytes. GSDME expression enhances the phagocytosis of tumour cells by tumour-associated macrophages, as well as the number and functions of tumour-infiltrating natural-killer and CD8+ T lymphocytes. Killer-cell granzyme B also activates caspase-independent pyroptosis in target cells by directly cleaving GSDME at the same site as caspase 3. Uncleavable or pore-defective GSDME proteins are not tumour suppressive. Thus, tumour GSDME acts as a tumour suppressor by activating pyroptosis, enhancing anti-tumour immunity.
Conflict of interest statement
Figures




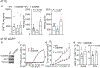




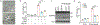


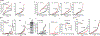


Comment in
-
Pyroptosis heats tumour immunity.Nat Rev Immunol. 2020 May;20(5):274-275. doi: 10.1038/s41577-020-0297-2. Nat Rev Immunol. 2020. PMID: 32203327 No abstract available.
-
Pyroptosis heats tumour immunity.Nat Rev Drug Discov. 2020 May;19(5):309. doi: 10.1038/d41573-020-00062-8. Nat Rev Drug Discov. 2020. PMID: 32251376 No abstract available.
-
Gasdermins deliver a deadly punch to cancer.Cell Res. 2020 Jun;30(6):463-464. doi: 10.1038/s41422-020-0316-7. Cell Res. 2020. PMID: 32327717 Free PMC article. No abstract available.
-
Gasdermin: a novel therapeutic target for tumour treatment by activating anti-tumour immunity.Signal Transduct Target Ther. 2020 May 13;5(1):69. doi: 10.1038/s41392-020-0180-4. Signal Transduct Target Ther. 2020. PMID: 32404864 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

