Genes regulating hormone stimulus and response to protein signaling revealed differential expression pattern during porcine oocyte in vitro maturation, confirmed by lipid concentration
- PMID: 32189110
- PMCID: PMC7343741
- DOI: 10.1007/s00418-020-01866-w
Genes regulating hormone stimulus and response to protein signaling revealed differential expression pattern during porcine oocyte in vitro maturation, confirmed by lipid concentration
Abstract
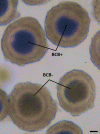
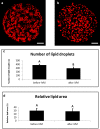
Genes influencing oocyte maturation may be valuable for predicting their developmental potential, as well as discerning the mechanistic pathways regulating oocyte development. In the presented research microarray gene expression analysis of immature and in vitro matured porcine oocytes was performed. Two groups of oocytes were compared in the study: before (3 × n = 50) and after in vitro maturation (3 × n = 50). The selection of viable oocytes was performed using the brilliant cresyl blue (BCB) test. Furthermore, microarrays and RT-qPCR was used to analyze the transcriptome of the oocytes before and after IVM. The study focused on the genes undergoing differential expression in two gene-ontology groups: "Cellular response to hormone stimulus" and "Cellular response to unfolded protein", which contain genes that may directly or indirectly be involved in signal transduction during oocyte maturation. Examination of all the genes of interest showed a lower level of their expression after IVM. From the total number of genes in these gene ontologies ten of the highest change in expression were identified: FOS, ID2, BTG2, CYR61, ESR1, AR, TACR3, CCND2, EGR2 and TGFBR3. The successful maturation of the oocytes was additionally confirmed with the use of lipid droplet assay. The genes were briefly described and related to the literature sources, to investigate their potential roles in the process of oocyte maturation. The results of the study may serve as a basic molecular reference for further research aimed at improving the methods of oocyte in vitro maturation, which plays an important role in the procedures of assisted reproduction.
Keywords: Microarray; Mitochondrial activity; Oocyte maturation; Pig.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





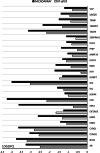
References
-
- Artini PG, Tatone C, Sperduti S, D’Aurora M, Franchi S, Di Emidio G, Gatta V, et al. Cumulus cells surrounding oocytes with high developmental competence exhibit down-regulation of phosphoinositol 1, 3 kinase/protein kinase B (PI3K/AKT) signalling genes involved in proliferation and survival. Hum Reprod. 2017;32(12):2474–2484. doi: 10.1093/humrep/dex320. - DOI - PMC - PubMed
-
- Blaha M, Nemcova L, Kepkova KV, Vodicka P, Prochazka R. Gene expression analysis of pig cumulus-oocyte complexes stimulated in vitro with follicle stimulating hormone or epidermal growth factor-like peptides. Reprod Biol Endocrinol. 2015;13(1):113. doi: 10.1186/s12958-015-0112-2. - DOI - PMC - PubMed
-
- Borys-Wójcik S, Kocherova I, Celichowski P, Popis M, Jeseta M, Bukowska D, Kempisty B, et al. Protein oligomerization is the biochemical process highly up-regulated in porcine oocytes before in vitro maturation (IVM) Med J Cell Biol. 2018;6(4):155–162. doi: 10.2478/acb-2018-0025. - DOI
-
- Borys S, Brązert M, Jankowski M, Kocherova I, Ożegowska K, Celichowski P, Kempisty B, et al. Enzyme linked receptor protein signaling pathway is one of the ontology groups that are highly up-regulated in porcine oocytes before in vitro maturation. J Biol Regul Homeost Agents. 2018;32(5):21–35. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

