Regulatory circuits controlling Spx levels in Streptococcus mutans
- PMID: 32189382
- PMCID: PMC7367440
- DOI: 10.1111/mmi.14499
Regulatory circuits controlling Spx levels in Streptococcus mutans
Abstract
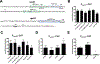
Spx is a major regulator of stress responses in Firmicutes. In Streptococcus mutans, two Spx homologues, SpxA1 and SpxA2, were identified as mediators of oxidative stress responses but the regulatory circuits controlling their levels and activity are presently unknown. Comparison of SpxA1 and SpxA2 protein sequences revealed differences at the C-terminal end, with SpxA1 containing an unusual number of acidic residues. Here, we showed that a green fluorescence protein (GFP) reporter becomes unstable when fused to the last 10 amino acids of SpxA2 but remained stable when fused to the C-terminal acidic tail of SpxA1. Inactivation of clpP or simultaneous inactivation of clpC and clpE stabilized the GFP::SpxA2tail fusion protein. Addition of acidic amino acids to the GFP::SpxA2tail chimera stabilized GFP, while deletion of the acidic residues destabilized GFP::SpxA1tail . Promoter reporter fusions revealed that spxA1 transcription is co-repressed by the metalloregulators PerR and SloR while spxA2 transcription is largely dependent on the envelope stress regulator LiaFSR. In agreement with spxA2 being part of the LiaR regulon, SpxA2 was found to be critical for the growth of S. mutans under envelope stress conditions. Finally, we showed that redox sensing is essential for SpxA1-dependent activation of oxidative stress responses but dispensable for SpxA2-mediated envelope stress responses.
Keywords: Streptococcus mutans; ClpP; Spx; oxidative stress.
© 2020 John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest.
Figures


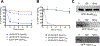


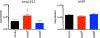
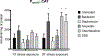


Similar articles
-
Transcription of Oxidative Stress Genes Is Directly Activated by SpxA1 and, to a Lesser Extent, by SpxA2 in Streptococcus mutans.J Bacteriol. 2015 Jul;197(13):2160-2170. doi: 10.1128/JB.00118-15. Epub 2015 Apr 20. J Bacteriol. 2015. PMID: 25897032 Free PMC article.
-
Inactivation of the spxA1 or spxA2 gene of Streptococcus mutans decreases virulence in the rat caries model.Mol Oral Microbiol. 2017 Apr;32(2):142-153. doi: 10.1111/omi.12160. Epub 2016 May 16. Mol Oral Microbiol. 2017. PMID: 27037617 Free PMC article.
-
Transcriptional and Phenotypic Characterization of Novel Spx-Regulated Genes in Streptococcus mutans.PLoS One. 2015 Apr 23;10(4):e0124969. doi: 10.1371/journal.pone.0124969. eCollection 2015. PLoS One. 2015. PMID: 25905865 Free PMC article.
-
Streptococcus mutans SpxA2 relays the signal of cell envelope stress from LiaR to effectors that maintain cell wall and membrane homeostasis.Mol Oral Microbiol. 2020 Jun;35(3):118-128. doi: 10.1111/omi.12282. Epub 2020 Feb 26. Mol Oral Microbiol. 2020. PMID: 32043713 Free PMC article.
-
Molecular and regulatory mechanisms of oxidative stress adaptation in Streptococcus mutans.Mol Oral Microbiol. 2023 Feb;38(1):1-8. doi: 10.1111/omi.12388. Epub 2022 Sep 25. Mol Oral Microbiol. 2023. PMID: 36088636 Review.
Cited by
-
Natural compounds: new therapeutic approach for inhibition of Streptococcus mutans and dental caries.Front Pharmacol. 2025 Apr 1;16:1548117. doi: 10.3389/fphar.2025.1548117. eCollection 2025. Front Pharmacol. 2025. PMID: 40235544 Free PMC article. Review.
-
A Genome-Wide CRISPR Interference Screen Reveals an StkP-Mediated Connection between Cell Wall Integrity and Competence in Streptococcus salivarius.mSystems. 2022 Dec 20;7(6):e0073522. doi: 10.1128/msystems.00735-22. Epub 2022 Nov 7. mSystems. 2022. PMID: 36342134 Free PMC article.
-
Deciphering the role of SMU.1147 in peptide-mediated signaling and competence in Streptococcus mutans.Microbiol Spectr. 2025 Apr;13(4):e0291724. doi: 10.1128/spectrum.02917-24. Epub 2025 Mar 5. Microbiol Spectr. 2025. PMID: 40042332 Free PMC article.
-
ClpX/P-Dependent Degradation of Novel Substrates in Streptococcus mutans.J Bacteriol. 2022 Apr 19;204(4):e0059421. doi: 10.1128/jb.00594-21. Epub 2022 Mar 28. J Bacteriol. 2022. PMID: 35343773 Free PMC article.
-
LiaR-dependent gene expression contributes to antimicrobial responses in group A Streptococcus.Antimicrob Agents Chemother. 2024 Dec 5;68(12):e0049624. doi: 10.1128/aac.00496-24. Epub 2024 Nov 13. Antimicrob Agents Chemother. 2024. PMID: 39535201 Free PMC article.
References
-
- Baek KT, Thogersen L, Mogenssen RG, Mellergaard M, Thomsen LE, Petersen A, … Frees D (2015). Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob Agents Chemother, 59(11), 6983–6991. 10.1128/AAC.01303-15 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

