A Protein Corona Adsorbed to a Bacterial Magnetosome Affects Its Cellular Uptake
- PMID: 32189964
- PMCID: PMC7065717
- DOI: 10.2147/IJN.S220082
A Protein Corona Adsorbed to a Bacterial Magnetosome Affects Its Cellular Uptake
Abstract
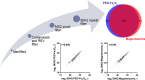
Purpose: It is well known that when exposed to human blood plasma, nanoparticles are predominantly coated by a layer of proteins, forming a corona that will mediate the subsequent cell interactions. Magnetosomes are protein-rich membrane nanoparticles which are synthesized by magnetic bacteria; these have gained a lot of attention owing to their unique magnetic and biochemical characteristics. Nevertheless, whether bacterial magnetosomes have a corona after interacting with the plasma, and how such a corona affects nanoparticle-cell interactions is yet to be elucidated. The aim of this study was to characterize corona formation around a bacterial magnetosome and to assess the functional consequences.
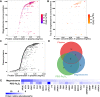
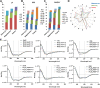
Methods: Magnetosomes were isolated from the magnetotactic bacteria, M. gryphiswaldense (MSR-1). Size, morphology, and zeta potential were measured by transmission electron microscopy and dynamic light scattering. A quantitative characterization of plasma corona proteins was performed using LC-MS/MS. Protein absorption was further examined by circular dichroism and the effect of the corona on cellular uptake was investigated by microscopy and spectroscopy.
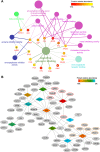
Results: Various serum proteins were found to be selectively adsorbed on the surface of the bacterial magnetosomes following plasma exposure, forming a corona. Compared to the pristine magnetosomes, the acquired corona promoted efficient cellular uptake by human vascular endothelial cells. Using a protein-interaction prediction method, we identified cell surface receptors that could potentially associate with abundant corona components. Of these, one abundant corona protein, ApoE, may be responsible for internalization of the magnetosome-corona complex through LDL receptor-mediated internalization.
Conclusion: Our findings provide clues as to the physiological response to magnetosomes and also reveal the corona composition of this membrane-coated nanomaterial after exposure to blood plasma.
Keywords: LC-MS/MS; biogenic magnetic nanoparticle; cellular interaction; cellular uptake.
© 2020 Lai et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures

References
-
- Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome Island which undergoes frequent rearrangements during stationary growth. J Bacteriol. 2005;187(21):7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005 - DOI - PMC - PubMed
-
- Bazylinski DA, Lefèvre CT, Lower BH. Magnetotactic bacteria, magnetosomes, and nanotechnology In: Barton LL, Bazylinski DA, Xu H, editors. Nanomicrobiology: Physiological and Environmental Characteristics. New York (NY): Springer; 2014:39–74.
-
- Matsunaga T, Arakaki A. Molecular bioengineering of bacterial magnetic particles for biotechnological applications In: Schüler D, editor. Magnetoreception and Magnetosomes in Bacteria. Berlin, Heidelberg: Springer; 2007:227–254.
-
- Komeili A. Cell biology of magnetosome formation In: Schüler D, editor. Magnetoreception and Magnetosomes in Bacteria. Berlin, Heidelberg: Springer; 2007:163–174.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

