Biosynthesis of plant tetrahydroisoquinoline alkaloids through an imine reductase route
- PMID: 32190259
- PMCID: PMC7067268
- DOI: 10.1039/c9sc03773j
Biosynthesis of plant tetrahydroisoquinoline alkaloids through an imine reductase route
Abstract
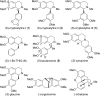
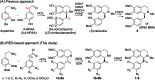
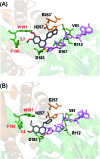
Herein, we report a biocatalytic approach to synthesize plant tetrahydroisoquinoline alkaloids (THIQAs) from dihydroisoquinoline (DHIQ) precursors using imine reductases and N-methyltransferase (NMT). The imine reductase IR45 was engineered to significantly expand its substrate specificity, enabling efficient and stereoselective conversion of 1-phenyl and 1-benzyl 6,7-dimethoxy-DHIQs into the corresponding (S)-tetrahydroisoquinolines (S-THIQs). Coclaurine N-methyltransferase (CNMT) was able to further efficiently convert these (S)-THIQ intermediates into (S)-THIQAs. By assembling IRED, CNMT, and glucose dehydrogenase (GDH) in one reaction, we effectively constituted two artificial biosynthetic pathways in Escherichia coli and successfully applied them to the production of five (S)-THIQAs. This highly efficient (100% yield from DHIQs) and easily tailorable (adding other genes) biosynthetic approach will be useful for producing a variety of plant THIQAs.
This journal is © The Royal Society of Chemistry 2020.
Figures


Similar articles
-
Asymmetric Synthesis of Fused-Ring Tetrahydroisoquinolines and Tetrahydro-β-carbolines from 2-Arylethylamines via a Chemoenzymatic Approach.Org Lett. 2022 Sep 16;24(36):6531-6536. doi: 10.1021/acs.orglett.2c02466. Epub 2022 Sep 6. Org Lett. 2022. PMID: 36066397
-
Introduction of chirality at C1 position of 1-substituted-3,4-dihydroisoquinoline by its enantioselective reduction: synthesis of chiral 1-substituted-1,2,3,4-tetrahydroisoquinoline - a review.RSC Adv. 2023 Apr 6;13(16):11010-11036. doi: 10.1039/d3ra01413d. eCollection 2023 Apr 3. RSC Adv. 2023. PMID: 37033430 Free PMC article. Review.
-
The Discovery of Imine Reductases and their Utilisation for the Synthesis of Tetrahydroisoquinolines.ChemCatChem. 2023 Feb 8;15(3):e202201126. doi: 10.1002/cctc.202201126. Epub 2023 Jan 11. ChemCatChem. 2023. PMID: 37081856 Free PMC article.
-
A single residue determines substrate preference in benzylisoquinoline alkaloid N-methyltransferases.Phytochemistry. 2020 Feb;170:112193. doi: 10.1016/j.phytochem.2019.112193. Epub 2019 Nov 23. Phytochemistry. 2020. PMID: 31765874
-
Biosynthesis of Tetrahydroisoquinoline Antibiotics.Curr Top Med Chem. 2016;16(15):1717-26. doi: 10.2174/1568026616666151012112329. Curr Top Med Chem. 2016. PMID: 26456466 Review.
Cited by
-
Computational Study of Mechanism and Enantioselectivity of Imine Reductase from Amycolatopsis orientalis.ChemistryOpen. 2022 Jan;11(1):e202100250. doi: 10.1002/open.202100250. Epub 2021 Nov 25. ChemistryOpen. 2022. PMID: 34825518 Free PMC article.
-
The role of biocatalysis in the asymmetric synthesis of alkaloids - an update.RSC Adv. 2021 Aug 20;11(45):28223-28270. doi: 10.1039/d1ra04181a. eCollection 2021 Aug 16. RSC Adv. 2021. PMID: 35480754 Free PMC article. Review.
-
Synthetic Biology in Plants, a Boon for Coming Decades.Mol Biotechnol. 2021 Dec;63(12):1138-1154. doi: 10.1007/s12033-021-00386-9. Epub 2021 Aug 21. Mol Biotechnol. 2021. PMID: 34420149 Review.
-
Recent trends in biocatalysis.Chem Soc Rev. 2021 Jul 21;50(14):8003-8049. doi: 10.1039/d0cs01575j. Epub 2021 Jun 18. Chem Soc Rev. 2021. PMID: 34142684 Free PMC article. Review.
-
Engineering a norcoclaurine synthase for one-step synthesis of (S)-1-aryl-tetrahydroisoquinolines.Bioresour Bioprocess. 2023 Mar 1;10(1):15. doi: 10.1186/s40643-023-00637-4. Bioresour Bioprocess. 2023. PMID: 38647611 Free PMC article.
References
-
- Liu W., Liu S., Jin R., Guo H., Zhao J. Org. Chem. Front. 2015;2:288–299.
- Chrzanowska M., Grajewska A., Rozwadowska M. D. Chem. Rev. 2016;116:12369–12465. - PubMed
-
- Perez M., Wu Z., Scalone M., Ayad T., Ratovelomanana-Vidal V. Eur. J. Org. Chem. 2015:6503–6514.
- Singh I. P., Shah P. Expert Opin. Ther. Pat. 2017;27:17–36. - PubMed
-
- Narcross L., Fossati E., Bourgeois L., Dueber J. E., Martin V. J. J. Trends Biotechnol. 2016;34:228–241. - PubMed
-
- Stadler R., Kutchan T. M., Zenk M. H. Phytochemistry. 1989;28:1083–1086.
- De-Eknamkul W., Suttipanta N., Kutchan T. M. Phytochemistry. 2000;55:177–181. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

