Identification of potential key genes in gastric cancer using bioinformatics analysis
- PMID: 32190306
- PMCID: PMC7054703
- DOI: 10.3892/br.2020.1281
Identification of potential key genes in gastric cancer using bioinformatics analysis
Abstract
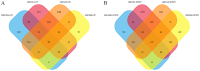
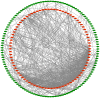
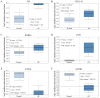
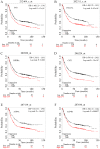
Gastric cancer (GC) is one of the most common types of cancer worldwide. Patients must be identified at an early stage of tumor progression for treatment to be effective. The aim of the present study was to identify potential biomarkers with diagnostic value in patients with GC. To examine potential therapeutic targets for GC, four Gene Expression Omnibus (GEO) datasets were downloaded and screened for differentially expressed genes (DEGs). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were subsequently performed to study the function and pathway enrichment of the identified DEGs. A protein-protein interaction (PPI) network was constructed. The CytoHubba plugin of Cytoscape was used to calculate the degree of connectivity of proteins in the PPI network, and the two genes with the highest degree of connectivity were selected for further analysis. Additionally, the two DEGs with the largest and smallest log Fold Change values were selected. These six key genes were further examined using Oncomine and the Kaplan-Meier plotter platform. A total of 99 upregulated and 172 downregulated genes common to all four GEO datasets were screened. The DEGs were primarily enriched in the Biological Process terms: 'extracellular matrix organization', 'collagen catabolic process' and 'cell adhesion'. These three KEGG pathways were significantly enriched in the categories: 'ECM-receptor interaction', 'protein digestion and absorption', and 'focal adhesion'. Based on Oncomine, expression of ATP4A and ATP4B were downregulated in GC, whereas expression of the other genes were all upregulated. The Kaplan-Meier plotter platform confirmed that upregulated expression of the identified key genes was significantly associated with worse overall survival of patients with GC. The results of the present study suggest that FN1, COL1A1, INHBA and CST1 may be potential biomarkers and therapeutic targets for GC. Additional studies are required to explore the potential value of ATP4A and ATP4B in the treatment of GC.
Keywords: bioinformatics analysis; diagnosis; differentially expressed genes; gastric cancer; key genes.
Copyright: © Wang et al.
Figures



Similar articles
-
Identification of hub genes and potential molecular mechanisms in gastric cancer by integrated bioinformatics analysis.PeerJ. 2018 Jul 2;6:e5180. doi: 10.7717/peerj.5180. eCollection 2018. PeerJ. 2018. PMID: 30002985 Free PMC article.
-
In silico analyses for potential key genes associated with gastric cancer.PeerJ. 2018 Dec 7;6:e6092. doi: 10.7717/peerj.6092. eCollection 2018. PeerJ. 2018. PMID: 30568862 Free PMC article.
-
Identification of key pathways and genes in nasopharyngeal carcinoma using bioinformatics analysis.Oncol Lett. 2019 May;17(5):4683-4694. doi: 10.3892/ol.2019.10133. Epub 2019 Mar 8. Oncol Lett. 2019. PMID: 30988824 Free PMC article.
-
Identification of candidate biomarkers and pathways associated with SCLC by bioinformatics analysis.Mol Med Rep. 2018 Aug;18(2):1538-1550. doi: 10.3892/mmr.2018.9095. Epub 2018 May 29. Mol Med Rep. 2018. PMID: 29845250 Free PMC article.
-
Collagen family genes and related genes might be associated with prognosis of patients with gastric cancer: an integrated bioinformatics analysis and experimental validation.Transl Cancer Res. 2020 Oct;9(10):6246-6262. doi: 10.21037/tcr-20-1726. Transl Cancer Res. 2020. PMID: 35117235 Free PMC article.
Cited by
-
Identification of Potential Diagnostic and Prognostic Biomarkers for Gastric Cancer Based on Bioinformatic Analysis.Front Genet. 2022 Mar 16;13:862105. doi: 10.3389/fgene.2022.862105. eCollection 2022. Front Genet. 2022. PMID: 35368700 Free PMC article.
-
CGB5, INHBA and TRAJ19 Hold Prognostic Potential as Immune Genes for Patients with Gastric Cancer.Dig Dis Sci. 2023 Mar;68(3):791-802. doi: 10.1007/s10620-022-07513-9. Epub 2022 May 27. Dig Dis Sci. 2023. PMID: 35624327
-
α-Enolase Lies Downstream of mTOR/HIF1α and Promotes Thyroid Carcinoma Progression by Regulating CST1.Front Cell Dev Biol. 2021 Apr 21;9:670019. doi: 10.3389/fcell.2021.670019. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33968941 Free PMC article.
-
Identification of Differentially Expressed Genes Reveals BGN Predicting Overall Survival and Tumor Immune Infiltration of Gastric Cancer.Comput Math Methods Med. 2021 Nov 26;2021:5494840. doi: 10.1155/2021/5494840. eCollection 2021. Comput Math Methods Med. 2021. PMID: 34868341 Free PMC article.
-
Identification of gene profiles related to the development of oral cancer using a deep learning technique.BMC Med Genomics. 2023 Feb 27;16(1):35. doi: 10.1186/s12920-023-01462-6. BMC Med Genomics. 2023. PMID: 36849997 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
