Longitudinal study reveals HIV-1-infected CD4+ T cell dynamics during long-term antiretroviral therapy
- PMID: 32191639
- PMCID: PMC7324206
- DOI: 10.1172/JCI135953
Longitudinal study reveals HIV-1-infected CD4+ T cell dynamics during long-term antiretroviral therapy
Abstract
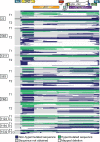
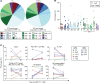
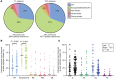
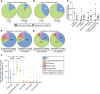
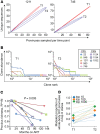
Proliferation of CD4+ T cells harboring HIV-1 proviruses is a major contributor to viral persistence in people on antiretroviral therapy (ART). To determine whether differential rates of clonal proliferation or HIV-1-specific cytotoxic T lymphocyte (CTL) pressure shape the provirus landscape, we performed an intact proviral DNA assay (IPDA) and obtained 661 near-full-length provirus sequences from 8 individuals with suppressed viral loads on ART at time points 7 years apart. We observed slow decay of intact proviruses but no changes in the proportions of various types of defective proviruses. The proportion of intact proviruses in expanded clones was similar to that of defective proviruses in clones. Intact proviruses observed in clones did not have more escaped CTL epitopes than intact proviruses observed as singlets. Concordantly, total proviruses at later time points or observed in clones were not enriched in escaped or unrecognized epitopes. Three individuals with natural control of HIV-1 infection (controllers) on ART, included because controllers have strong HIV-1-specific CTL responses, had a smaller proportion of intact proviruses but a distribution of defective provirus types and escaped or unrecognized epitopes similar to that of the other individuals. This work suggests that CTL selection does not significantly check clonal proliferation of infected cells or greatly alter the provirus landscape in people on ART.
Keywords: AIDS/HIV; Adaptive immunity; Antigen presentation; T cells.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- UM1 AI126620/AI/NIAID NIH HHS/United States
- K25 AI155224/AI/NIAID NIH HHS/United States
- R01 AI158013/AI/NIAID NIH HHS/United States
- T32 AI007291/AI/NIAID NIH HHS/United States
- P30 AI094189/AI/NIAID NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- UM1 AI126623/AI/NIAID NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- P30 AI027757/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- UM1 AI126603/AI/NIAID NIH HHS/United States
- UM1 AI126611/AI/NIAID NIH HHS/United States
- K08 AI143391/AI/NIAID NIH HHS/United States

