Lexis and Grammar of Mitochondrial RNA Processing in Trypanosomes
- PMID: 32191849
- PMCID: PMC7083771
- DOI: 10.1016/j.pt.2020.01.006
Lexis and Grammar of Mitochondrial RNA Processing in Trypanosomes
Abstract
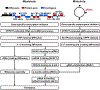
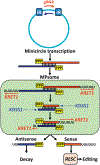
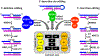
Trypanosoma brucei spp. cause African human and animal trypanosomiasis, a burden on health and economy in Africa. These hemoflagellates are distinguished by a kinetoplast nucleoid containing mitochondrial DNAs of two kinds: maxicircles encoding ribosomal RNAs (rRNAs) and proteins and minicircles bearing guide RNAs (gRNAs) for mRNA editing. All RNAs are produced by a phage-type RNA polymerase as 3' extended precursors, which undergo exonucleolytic trimming. Most pre-mRNAs proceed through 3' adenylation, uridine insertion/deletion editing, and 3' A/U-tailing. The rRNAs and gRNAs are 3' uridylated. Historically, RNA editing has attracted major research effort, and recently essential pre- and postediting processing events have been discovered. Here, we classify the key players that transform primary transcripts into mature molecules and regulate their function and turnover.
Keywords: RNA decay; RNA editing; Trypanosoma; kinetoplast; mitochondria; polyadenylation.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures

References
-
- Maslov DA et al. (2019) Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology 146, 1–27 - PubMed
-
- Ramrath DJF et al. (2018) Evolutionary shift toward protein based architecture in trypanosomal mitochondrial ribosomes. Science 362, eaau7735. - PubMed
-
- Hancock K and Hajduk SL (1990) The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem 265, 19208–19215 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM084065/GM/NIGMS NIH HHS/United States
- R01 AI091914/AI/NIAID NIH HHS/United States
- R01 GM129041/GM/NIGMS NIH HHS/United States
- R01 AI014102/AI/NIAID NIH HHS/United States
- R01 AI064653/AI/NIAID NIH HHS/United States
- R15 AI135885/AI/NIAID NIH HHS/United States
- R01 AI061580/AI/NIAID NIH HHS/United States
- R21 AI139448/AI/NIAID NIH HHS/United States
- R01 AI113157/AI/NIAID NIH HHS/United States
- R01 AI101057/AI/NIAID NIH HHS/United States
- R01 AI125487/AI/NIAID NIH HHS/United States
- MR/L019701/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous

