FADD Phosphorylation Modulates Blood Glucose Levels by Decreasing the Expression of Insulin-Degrading Enzyme
- PMID: 32191993
- PMCID: PMC7191044
- DOI: 10.14348/molcells.2020.2198
FADD Phosphorylation Modulates Blood Glucose Levels by Decreasing the Expression of Insulin-Degrading Enzyme
Abstract
Our previous study revealed a novel role of Fas-associated death domain-containing protein (FADD) in islet development and insulin secretion. Insulin-degrading enzyme (IDE) is a zinc metalloprotease that selectively degrades biologically important substrates associated with type 2 diabetes (T2DM). The current study was designed to investigate the effect of FADD phosphorylation on IDE. We found that the mRNA and protein levels of IDE were significantly downregulated in FADD-D mouse livers compared with control mice. Quantitative real-time polymerase chain reaction analysis showed that FADD regulates the expression of IDE at the transcriptional level without affecting the stability of the mRNA in HepG2 cells. Following treatment with cycloheximide, the IDE protein degradation rate was found to be increased in both FADD-D primary hepatocytes and FADD-knockdown HepG2 cells. Additionally, IDE expression levels were reduced in insulin-stimulated primary hepatocytes from FADD-D mice compared to those from control mice. Moreover, FADD phosphorylation promotes nuclear translocation of FoxO1, thus inhibiting the transcriptional activity of the IDE promoter. Together, these findings imply a novel role of FADD in the reduction of protein stability and expression levels of IDE.
Keywords: FADD phosphorylation; FoxO1; insulin; insulin-degrading enzyme; protein stability.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures

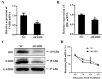

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

