Genome-Wide Association Mapping for Heat Stress Responsive Traits in Field Pea
- PMID: 32192061
- PMCID: PMC7139655
- DOI: 10.3390/ijms21062043
Genome-Wide Association Mapping for Heat Stress Responsive Traits in Field Pea
Abstract
Environmental stress hampers pea productivity. To understand the genetic basis of heat resistance, a genome-wide association study (GWAS) was conducted on six stress responsive traits of physiological and agronomic importance in pea, with an objective to identify the genetic loci associated with these traits. One hundred and thirty-five genetically diverse pea accessions from major pea growing areas of the world were phenotyped in field trials across five environments, under generally ambient (control) and heat stress conditions. Statistical analysis of phenotype indicated significant effects of genotype (G), environment (E), and G × E interaction for all traits. A total of 16,877 known high-quality SNPs were used for association analysis to determine marker-trait associations (MTA). We identified 32 MTAs that were consistent in at least three environments for association with the traits of stress resistance: six for chlorophyll concentration measured by a soil plant analysis development meter; two each for photochemical reflectance index and canopy temperature; seven for reproductive stem length; six for internode length; and nine for pod number. Forty-eight candidate genes were identified within 15 kb distance of these markers. The identified markers and candidate genes have potential for marker-assisted selection towards the development of heat resistant pea cultivars.
Keywords: GWAS; candidate-gene; genetic diversity; genotyping-by-sequencing; heat stress; marker-trait association; pea.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

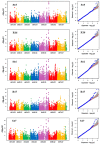
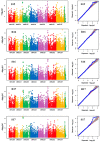
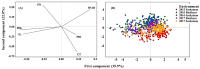
Similar articles
-
Genome-Wide Association Mapping for Heat and Drought Adaptive Traits in Pea.Genes (Basel). 2021 Nov 26;12(12):1897. doi: 10.3390/genes12121897. Genes (Basel). 2021. PMID: 34946846 Free PMC article.
-
Genetic dissection of heat-responsive physiological traits to improve adaptation and increase yield potential in soft winter wheat.BMC Genomics. 2020 Apr 20;21(1):315. doi: 10.1186/s12864-020-6717-7. BMC Genomics. 2020. PMID: 32312234 Free PMC article.
-
Genome-wide association study identifies favorable SNP alleles and candidate genes for frost tolerance in pea.BMC Genomics. 2020 Aug 4;21(1):536. doi: 10.1186/s12864-020-06928-w. BMC Genomics. 2020. PMID: 32753054 Free PMC article.
-
Omics resources and omics-enabled approaches for achieving high productivity and improved quality in pea (Pisum sativum L.).Theor Appl Genet. 2021 Mar;134(3):755-776. doi: 10.1007/s00122-020-03751-5. Epub 2021 Jan 12. Theor Appl Genet. 2021. PMID: 33433637 Review.
-
Perspectives on the genetic improvement of health- and nutrition-related traits in pea.Plant Physiol Biochem. 2021 Jan;158:353-362. doi: 10.1016/j.plaphy.2020.11.020. Epub 2020 Nov 17. Plant Physiol Biochem. 2021. PMID: 33250319 Free PMC article. Review.
Cited by
-
QTL mapping and underlying genes for heat tolerance in grapevine (Rhine Riesling × Cabernet Sauvignon) under field conditions.Theor Appl Genet. 2025 Jul 21;138(8):189. doi: 10.1007/s00122-025-04972-2. Theor Appl Genet. 2025. PMID: 40691321 Free PMC article.
-
Stable SNP Allele Associations With High Grain Zinc Content in Polished Rice (Oryza sativa L.) Identified Based on ddRAD Sequencing.Front Genet. 2020 Aug 11;11:763. doi: 10.3389/fgene.2020.00763. eCollection 2020. Front Genet. 2020. PMID: 32849786 Free PMC article.
-
Genome-wide association study for morphological traits and resistance to Peryonella pinodes in the USDA pea single plant plus collection.G3 (Bethesda). 2022 Aug 25;12(9):jkac168. doi: 10.1093/g3journal/jkac168. G3 (Bethesda). 2022. PMID: 35792880 Free PMC article.
-
Analysis and Identification of QTL for Resistance to Sclerotinia sclerotiorum in Pea (Pisum sativum L.).Front Genet. 2020 Nov 19;11:587968. doi: 10.3389/fgene.2020.587968. eCollection 2020. Front Genet. 2020. PMID: 33329732 Free PMC article.
-
Physiological and Molecular Approaches for Developing Thermotolerance in Vegetable Crops: A Growth, Yield and Sustenance Perspective.Front Plant Sci. 2022 Jun 28;13:878498. doi: 10.3389/fpls.2022.878498. eCollection 2022. Front Plant Sci. 2022. PMID: 35837452 Free PMC article.
References
-
- Cousin R. Peas (Pisum sativum L.) Field Crop. Res. 1997;53:111–130. doi: 10.1016/S0378-4290(97)00026-9. - DOI
-
- Smýkal P., Aubert G., Burstin J., Coyne C.J., Ellis N.T.H., Flavell A.J., Ford R., Hýbl M., Macas J., Neumann P., et al. Pea (Pisum sativum L.) in the Genomic Era. Agronomy. 2012;2:74–115. doi: 10.3390/agronomy2020074. - DOI
-
- Guilioni L., Wery J., Tardieu F. Heat stress-induced abortion of buds and flowers in pea: Is sensitivity linked to organ age or to relations between reproductive organs? Ann. Bot. 1997;80:159–168. doi: 10.1006/anbo.1997.0425. - DOI
-
- Bueckert R.A., Wagenhoffer S., Hnatowich G., Warkentin T.D. Effect of heat and precipitation on pea yield and reproductive performance in the field. Can. J. Plant. Sci. 2015;95:629–639. doi: 10.4141/cjps-2014-342. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

