Distribution, organization an evolutionary history of La and LARPs in eukaryotes
- PMID: 32192383
- PMCID: PMC7928011
- DOI: 10.1080/15476286.2020.1739930
Distribution, organization an evolutionary history of La and LARPs in eukaryotes
Abstract
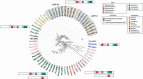
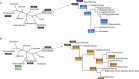
The fate of any cellular RNA is largely influenced by the nature and diversity of its interactions with various RNA-binding proteins (RBPs) leading to the formation of a biologically significant ribonucleoprotein (RNP) complex. La motif-containing proteins (composed of genuine La and La-related proteins (LARPs)) represent an evolutionary conserved family of RBPs that encompass a large range of crucial functions, involving coding and non-coding RNAs. In this work, we provide data that extend our previous knowledge on the distribution, organization and evolutionary history of this important protein family. Using a repertoire of 345 La motif-containing proteins from 135 species representing all major eukaryotic lineages, we were able to pinpoint many lineage-specific variations in the structural organization of La and LARPs and propose new evolutive scenarios to explain their modern genomic distribution.
Keywords: LARP; La-related proteins; RNA-binding proteins; evolution; genuine La.
Conflict of interest statement
No potential conflict of interest was reported by the author.
Figures





References
-
- Wolin SL, Cedervall T. The La protein. Annu Rev Biochem. 2002;71(1):375–403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
