Repositioning of Hypoglycemic Drug Linagliptin for Cancer Treatment
- PMID: 32194417
- PMCID: PMC7062795
- DOI: 10.3389/fphar.2020.00187
Repositioning of Hypoglycemic Drug Linagliptin for Cancer Treatment
Abstract
Background: Drug repositioning, development of new uses for marketed drugs, is an effective way to discover new antitumor compounds. In this study, we used a new method, filtering compounds via molecular docking to find key targets combination.
Methods: The data of gene expression in cancer and normal tissues of colorectal, breast, and liver cancer were obtained from The Cancer Genome Atlas Project (TCGA). The key targets combination was obtained from the protein-protein interaction network (PPI network) and the correlation analysis of the targets. Molecular docking was used to reposition the drugs which were obtained from DrugBank. MTT proliferation assay and animal experiments were used to verify the activity of candidate compounds. Flow cytometric analysis of proliferation, cell cycle and apoptosis, slice analysis, gene regulatory network, and Western blot were performed to elucidate the mechanism of drug action.
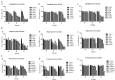
Results: CDK1 and AURKB were identified as a pair of key targets by the analysis of different expression gene from TCGA. Three compounds, linagliptin, mupirocin, and tobramycin, from 12 computationally predicted compounds, were verified to inhibit cell viability in HCT116 (colorectal), MCF7 (breast), and HepG2 (liver) cancer cells. Linagliptin, a hypoglycemic drug, was proved to inhibit cell proliferation by cell cycle arrest and induce apoptosis in HCT116 cells, and suppress tumor growth in nude mice bearing HCT116 cells. Linagliptin reduced the tumor size and decreased the expression of Ki67, a nuclear protein expressed in all proliferative cells. Gene regulatory network and Western blot analysis suggested that linagliptin inhibited tumor cell proliferation and promoted cell apoptosis through suppressing the expression and phosphorylation of Rb, plus down-regulating the expression of Pro-caspase3 and Bcl-2, respectively.
Conclusion: The combination of key targets based on the protein-protein interaction network that were built by the different gene expression of TCGA data to reposition the marketed drugs turned out to be a new approach to discover new antitumor drugs. Hypoglycemic drug linagliptin could potentially lead to novel therapeutics for the treatment of tumors, especially for colorectal cancer. Gene regulatory network is a valuable method for predicting and explaining the mechanism of drugs action.
Keywords: cell proliferation and apoptosis; drug repositioning; gene regulatory network; molecular docking; multi-target anti-tumor drug screening; the oncology genome atlas project; xenograft tumor mice.
Copyright © 2020 Li, Li, Li, Li, Quan, Wang and Sun.
Figures



Similar articles
-
The anti-tumor effect of OP-B on ovarian cancer in vitro and in vivo, and its mechanism: An investigation using network pharmacology-based analysis.J Ethnopharmacol. 2022 Jan 30;283:114706. doi: 10.1016/j.jep.2021.114706. Epub 2021 Oct 3. J Ethnopharmacol. 2022. PMID: 34614446
-
[Molecular mechanism of Spatholobi Caulis in treatment of ovarian cancer based on network pharmacology and experimental verification].Zhongguo Zhong Yao Za Zhi. 2022 Feb;47(3):786-795. doi: 10.19540/j.cnki.cjcmm.20211103.703. Zhongguo Zhong Yao Za Zhi. 2022. PMID: 35178962 Chinese.
-
Integrating network pharmacology deciphers the action mechanism of Zuojin capsule in suppressing colorectal cancer.Phytomedicine. 2022 Feb;96:153881. doi: 10.1016/j.phymed.2021.153881. Epub 2021 Dec 9. Phytomedicine. 2022. PMID: 34942456
-
Qizhen capsule inhibits colorectal cancer by inducing NAG-1/GDF15 expression that mediated via MAPK/ERK activation.J Ethnopharmacol. 2021 Jun 12;273:113964. doi: 10.1016/j.jep.2021.113964. Epub 2021 Feb 26. J Ethnopharmacol. 2021. PMID: 33640439
-
Network pharmacology and molecular docking reveal the mechanism of Scopoletin against non-small cell lung cancer.Life Sci. 2021 Apr 1;270:119105. doi: 10.1016/j.lfs.2021.119105. Epub 2021 Jan 23. Life Sci. 2021. PMID: 33497736
Cited by
-
Single-cell transcriptional signature-based drug repurposing and in vitro evaluation in colorectal cancer.BMC Cancer. 2024 Mar 25;24(1):371. doi: 10.1186/s12885-024-12142-8. BMC Cancer. 2024. PMID: 38528462 Free PMC article.
-
Synergistic Cytotoxic Impact of Linagliptin - Ciprofloxacin Combination on Cervical Cancer Cell Line: Insights into Targeting Heat Shock Protein 60.Asian Pac J Cancer Prev. 2025 Jun 1;26(6):2117-2128. doi: 10.31557/APJCP.2025.26.6.2117. Asian Pac J Cancer Prev. 2025. PMID: 40542774 Free PMC article.
-
Antiproliferative Impact of Linagliptin on the Cervical Cancer Cell Line.Asian Pac J Cancer Prev. 2024 Sep 1;25(9):3293-3300. doi: 10.31557/APJCP.2024.25.9.3293. Asian Pac J Cancer Prev. 2024. PMID: 39342609 Free PMC article.
-
Current trends and future prospects of drug repositioning in gastrointestinal oncology.Front Pharmacol. 2024 Jan 4;14:1329244. doi: 10.3389/fphar.2023.1329244. eCollection 2023. Front Pharmacol. 2024. PMID: 38239190 Free PMC article. Review.
-
Validating Cell Surface Proteases as Drug Targets for Cancer Therapy: What Do We Know, and Where Do We Go?Cancers (Basel). 2022 Jan 26;14(3):624. doi: 10.3390/cancers14030624. Cancers (Basel). 2022. PMID: 35158891 Free PMC article. Review.
References
-
- Ayoub B. M., Attia Y. M., Ahmed M. S. (2018). Structural re-positioning, in silico molecular modelling, oxidative degradation, and biological screening of linagliptin as adenosine 3 receptor (ADORA3) modulators targeting hepatocellular carcinoma. J. Enzyme Inhib. Med. Chem. 33 (1), 858–866. 10.1080/14756366.2018.1462801 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

