The Genetic Landscape of Dystrophin Mutations in Italy: A Nationwide Study
- PMID: 32194622
- PMCID: PMC7063120
- DOI: 10.3389/fgene.2020.00131
The Genetic Landscape of Dystrophin Mutations in Italy: A Nationwide Study
Abstract
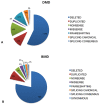
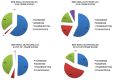
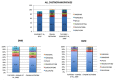
Dystrophinopathies are inherited diseases caused by mutations in the dystrophin (DMD) gene for which testing is mandatory for genetic diagnosis, reproductive choices and eligibility for personalized trials. We genotyped the DMD gene in our Italian cohort of 1902 patients (BMD n = 740, 39%; DMD n =1162, 61%) within a nationwide study involving 11 diagnostic centers in a 10-year window (2008-2017). In DMD patients, we found deletions in 57%, duplications in 11% and small mutations in 32%. In BMD, we found deletions in 78%, duplications in 9% and small mutations in 13%. In BMD, there are a higher number of deletions, and small mutations are more frequent than duplications. Among small mutations that are generally frequent in both phenotypes, 44% of DMD and 36% of BMD are nonsense, thus, eligible for stop codon read-through therapy; 63% of all out-of-frame deletions are eligible for single exon skipping. Patients were also assigned to Italian regions and showed interesting regional differences in mutation distribution. The full genetic characterization in this large, nationwide cohort has allowed us to draw several correlations between DMD/BMD genotype landscapes and mutation frequency, mutation types, mutation locations along the gene, exon/intron architecture, and relevant protein domain, with effects on population genetic characteristics and new personalized therapies.
Keywords: dystrophin; exon skipping therapy; muscular dystrophy; nationwide study; read-through therapy.
Copyright © 2020 Neri, Rossi, Trabanelli, Mauro, Selvatici, Falzarano, Spedicato, Margutti, Rimessi, Fortunato, Fabris, Gualandi, Comi, Tedeschi, Seia, Fiorillo, Traverso, Bruno, Giardina, Piemontese, Merla, Cau, Marica, Scuderi, Borgione, Tessa, Astrea, Santorelli, Merlini, Mora, Bernasconi, Gibertini, Sansone, Mongini, Berardinelli, Pini, Liguori, Filosto, Messina, Vita, Toscano, Vita, Pane, Servidei, Pegoraro, Bello, Travaglini, Bertini, D'Amico, Ergoli, Politano, Torella, Nigro, Mercuri and Ferlini.
Figures

Similar articles
-
Protein- and mRNA-based phenotype-genotype correlations in DMD/BMD with point mutations and molecular basis for BMD with nonsense and frameshift mutations in the DMD gene.Hum Mutat. 2007 Feb;28(2):183-95. doi: 10.1002/humu.20422. Hum Mutat. 2007. PMID: 17041906
-
DMD Mutations in 576 Dystrophinopathy Families: A Step Forward in Genotype-Phenotype Correlations.PLoS One. 2015 Aug 18;10(8):e0135189. doi: 10.1371/journal.pone.0135189. eCollection 2015. PLoS One. 2015. PMID: 26284620 Free PMC article.
-
Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase.Hum Mutat. 2009 Jun;30(6):934-45. doi: 10.1002/humu.20976. Hum Mutat. 2009. PMID: 19367636
-
MLPA Analyses Reveal a Spectrum of Dystrophin Gene Deletions/Duplications in Pakistani Patients Suspected of Having Duchenne/Becker Muscular Dystrophy: A Retrospective Study.Genet Test Mol Biomarkers. 2019 Jul;23(7):468-472. doi: 10.1089/gtmb.2018.0262. Epub 2019 May 31. Genet Test Mol Biomarkers. 2019. PMID: 31157985 Review.
-
Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations.Hum Mutat. 2009 Mar;30(3):293-9. doi: 10.1002/humu.20918. Hum Mutat. 2009. PMID: 19156838 Review.
Cited by
-
Biomarkers in Duchenne Muscular Dystrophy: Current Status and Future Directions.J Neuromuscul Dis. 2023;10(6):987-1002. doi: 10.3233/JND-221666. J Neuromuscul Dis. 2023. PMID: 37545256 Free PMC article. Review.
-
mRNA in situ hybridization exhibits unbalanced nuclear/cytoplasmic dystrophin transcript repartition in Duchenne myogenic cells and skeletal muscle biopsies.Sci Rep. 2023 Sep 24;13(1):15942. doi: 10.1038/s41598-023-43134-6. Sci Rep. 2023. PMID: 37743371 Free PMC article.
-
Theragnosis for Duchenne Muscular Dystrophy.Front Pharmacol. 2021 Jun 3;12:648390. doi: 10.3389/fphar.2021.648390. eCollection 2021. Front Pharmacol. 2021. PMID: 34149409 Free PMC article.
-
A Proof of Principle Proteomic Study Detects Dystrophin in Human Plasma: Implications in DMD Diagnosis and Clinical Monitoring.Int J Mol Sci. 2023 Mar 8;24(6):5215. doi: 10.3390/ijms24065215. Int J Mol Sci. 2023. PMID: 36982290 Free PMC article.
-
Genomic insights into Duchene muscular dystrophy: Analysis of 1250 patients reveals 30% novel genetic patterns and 6 novel variants.J Genet Eng Biotechnol. 2024 Dec;22(4):100436. doi: 10.1016/j.jgeb.2024.100436. Epub 2024 Nov 11. J Genet Eng Biotechnol. 2024. PMID: 39674649 Free PMC article.
References
-
- Aartsma-Rus A., Van Deutekom J. C., Fokkema I. F., Van Ommen G. J., Den Dunnen J. T. (2006). Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 34, 135–144. 10.1002/mus.20586 - DOI - PubMed
-
- Aartsma-Rus A., Straub V., Hemmings R., Haas M., Schlosser-Weber G., Stoyanova-Beninska V., et al. (2017). Development of exon skipping therapies for duchenne muscular dystrophy: a critical review and a perspective on the outstanding issues. Nucleic Acid Ther. 27, 251–259. 10.1007/s40265-016-0657-110.1089/nat.2017.0682 - DOI - PMC - PubMed

