Integrated Analysis of the Functions and Prognostic Values of RNA Binding Proteins in Lung Squamous Cell Carcinoma
- PMID: 32194639
- PMCID: PMC7066120
- DOI: 10.3389/fgene.2020.00185
Integrated Analysis of the Functions and Prognostic Values of RNA Binding Proteins in Lung Squamous Cell Carcinoma
Abstract
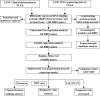
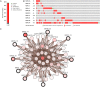
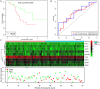
Lung cancer is the leading cause of cancer-related deaths worldwide. Dysregulation of RNA binding proteins (RBPs) has been found in a variety of cancers and is related to oncogenesis and progression. However, the functions of RBPs in lung squamous cell carcinoma (LUSC) remain unclear. In this study, we obtained gene expression data and corresponding clinical information for LUSC from The Cancer Genome Atlas (TCGA) database, identified aberrantly expressed RBPs between tumors and normal tissue, and conducted a series of bioinformatics analyses to explore the expression and prognostic value of these RBPs. A total of 300 aberrantly expressed RBPs were obtained, comprising 59 downregulated and 241 upregulated RBPs. Functional enrichment analysis indicated that the differentially expressed RBPs were mainly associated with mRNA metabolic processes, RNA processing, RNA modification, regulation of translation, the TGF-beta signaling pathway, and the Toll-like receptor signaling pathway. Nine RBP genes (A1CF, EIF2B5, LSM1, LSM7, MBNL2, RSRC1, TRMU, TTF2, and ZCCHC5) were identified as prognosis-associated hub genes by univariate, least absolute shrinkage and selection operator (LASSO), Kaplan-Meier survival, and multivariate Cox regression analyses, and were used to construct the prognostic model. Further analysis demonstrated that high risk scores for patients were significantly related to poor overall survival according to the model. The area under the time-dependent receiver operator characteristic curve of the prognostic model was 0.712 at 3 years and 0.696 at 5 years. We also developed a nomogram based on nine RBP genes, with internal validation in the TCGA cohort, which showed a favorable predictive efficacy for prognosis in LUSC. Our results provide novel insights into the pathogenesis of LUSC. The nine-RBP gene signature showed predictive value for LUSC prognosis, with potential applications in clinical decision-making and individualized treatment.
Keywords: RNA-binding proteins; bioinformatics; lung squamous cell carcinoma; prognostic signature; survival analysis.
Copyright © 2020 Li, Li, Gao and You.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources

