Regulation of cGAS-Mediated Immune Responses and Immunotherapy
- PMID: 32195086
- PMCID: PMC7080523
- DOI: 10.1002/advs.201902599
Regulation of cGAS-Mediated Immune Responses and Immunotherapy
Abstract
Early detection of infectious nucleic acids released from invading pathogens by the innate immune system is critical for immune defense. Detection of these nucleic acids by host immune sensors and regulation of DNA sensing pathways have been significant interests in the past years. Here, current understandings of evolutionarily conserved DNA sensing cyclic GMP-AMP (cGAMP) synthase (cGAS) are highlighted. Precise activation and tight regulation of cGAS are vital in appropriate innate immune responses, senescence, tumorigenesis and immunotherapy, and autoimmunity. Hence, substantial insights into cytosolic DNA sensing and immunotherapy of indispensable cytosolic sensors have been detailed to extend limited knowledge available thus far. This Review offers a critical, in-depth understanding of cGAS regulation, cytosolic DNA sensing, and currently established therapeutic approaches of essential cytosolic immune agents for improved human health.
Keywords: cGAS‐STING; cytosolic sensing; immunotherapy; innate immune regulation; tumorigenesis.
© 2020 The Authors. Published by WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
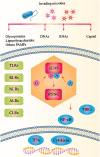
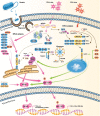
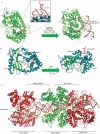
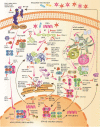

References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
