Microbiome-derived carnitine mimics as previously unknown mediators of gut-brain axis communication
- PMID: 32195337
- PMCID: PMC7065903
- DOI: 10.1126/sciadv.aax6328
Microbiome-derived carnitine mimics as previously unknown mediators of gut-brain axis communication
Abstract
Alterations to the gut microbiome are associated with various neurological diseases, yet evidence of causality and identity of microbiome-derived compounds that mediate gut-brain axis interaction remain elusive. Here, we identify two previously unknown bacterial metabolites 3-methyl-4-(trimethylammonio)butanoate and 4-(trimethylammonio)pentanoate, structural analogs of carnitine that are present in both gut and brain of specific pathogen-free mice but absent in germ-free mice. We demonstrate that these compounds are produced by anaerobic commensal bacteria from the family Lachnospiraceae (Clostridiales) family, colocalize with carnitine in brain white matter, and inhibit carnitine-mediated fatty acid oxidation in a murine cell culture model of central nervous system white matter. This is the first description of direct molecular inter-kingdom exchange between gut prokaryotes and mammalian brain cells, leading to inhibition of brain cell function.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution License 4.0 (CC BY).
Figures
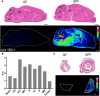
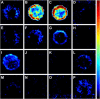
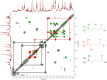

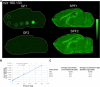

References
-
- Hsiao E. Y., McBride S. W., Hsien S., Sharon G., Hyde E. R., McCue T., Codelli J. A., Chow J., Reisman S. E., Petrosino J. F., Patterson P. H., Mazmanian S. K., Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

