An H3K9 methylation-dependent protein interaction regulates the non-enzymatic functions of a putative histone demethylase
- PMID: 32195666
- PMCID: PMC7192584
- DOI: 10.7554/eLife.53155
An H3K9 methylation-dependent protein interaction regulates the non-enzymatic functions of a putative histone demethylase
Abstract
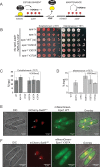
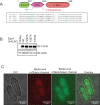
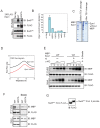
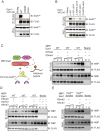
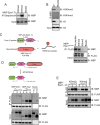
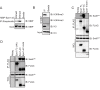
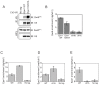
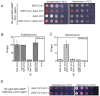
H3K9 methylation (H3K9me) specifies the establishment and maintenance of transcriptionally silent epigenetic states or heterochromatin. The enzymatic erasure of histone modifications is widely assumed to be the primary mechanism that reverses epigenetic silencing. Here, we reveal an inversion of this paradigm where a putative histone demethylase Epe1 in fission yeast, has a non-enzymatic function that opposes heterochromatin assembly. Mutations within the putative catalytic JmjC domain of Epe1 disrupt its interaction with Swi6HP1 suggesting that this domain might have other functions besides enzymatic activity. The C-terminus of Epe1 directly interacts with Swi6HP1, and H3K9 methylation stimulates this protein-protein interaction in vitro and in vivo. Expressing the Epe1 C-terminus is sufficient to disrupt heterochromatin by outcompeting the histone deacetylase, Clr3 from sites of heterochromatin formation. Our results underscore how histone modifying proteins that resemble enzymes have non-catalytic functions that regulate the assembly of epigenetic complexes in cells.
Keywords: S. pombe; biochemistry; chemical biology; chromosomes; demethylase; epigenetics; gene expression; heterochromatin; inheritance.
Plain language summary
A cell’s identity depends on which of its genes are active. One way for cells to control this process is to change how accessible their genes are to the molecular machinery that switches them on and off. Special proteins called histones determine how accessible genes are by altering how loosely or tightly DNA is packed together. Histones can be modified by enzymes, which are proteins that add or remove specific chemical ‘tags’. These tags regulate how accessible genes are and provide cells with a memory of gene activity. For example, a protein found in yeast called Epe1 helps reactivate large groups of genes after cell division, effectively ‘re-setting’ the yeast’s genome and eliminating past memories of the genes being inactive. For a long time, Epe1 was thought to do this by removing methyl groups, a ‘tag’ that indicates a gene is inactive, from histones – that is, by acting like an enzyme. However, no direct evidence to support this hypothesis has been found. Raiymbek et al. therefore set out to determine exactly how Epe1 worked, and whether or not it did indeed behave like an enzyme. Initial experiments testing mutant versions of Epe1 in yeast cells showed that the changes expected to stop Epe1 from removing methyl groups instead prevented the protein from ‘homing’ to the sections of DNA it normally activates. Detailed microscope imaging, using live yeast cells engineered to produce proteins with fluorescent markers, revealed that this inability to ‘home’ was due to a loss of interaction with Epe1’s main partner, a protein called Swi6. This protein recognizes and binds histones that have methyl tags. Swi6 also acts as a docking site for proteins involved in deactivating genes in close proximity to these histones. Further biochemical studies revealed how the interaction between Epe1 and Swi6 can help in gene reactivation. The methyl tag on histones in inactive regions of the genome inadvertently helps Epe1 interact more efficiently with Swi6. Then, Epe1 can simply block every other protein that binds to Swi6 from participating in gene deactivation. This observation contrasts with the prevailing view where the active removal of methyl tags by proteins such as Epe1 switches genes from an inactive to an active state. This work shows for the first time that Epe1 influences the state of the genome through a process that does not involve enzyme activity. In other words, although the protein may ‘moonlight’ as an enzyme, its main job uses a completely different mechanism. More broadly, these results increase the understanding of the many different ways that gene activity, and ultimately cell identity, can be controlled.
© 2020, Raiymbek et al.
Conflict of interest statement
GR, SA, NK, SG, AL, SB, RT, UC, KR No competing interests declared
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

