The origin of extracellular DNA in bacterial biofilm infections in vivo
- PMID: 32196074
- PMCID: PMC7150582
- DOI: 10.1093/femspd/ftaa018
The origin of extracellular DNA in bacterial biofilm infections in vivo
Abstract
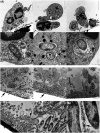
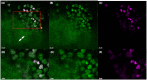
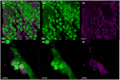
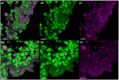
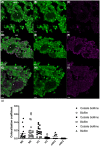
Extracellular DNA (eDNA) plays an important role in both the aggregation of bacteria and in the interaction of the resulting biofilms with polymorphonuclear leukocytes (PMNs) during an inflammatory response. Here, transmission electron and confocal scanning laser microscopy were used to examine the interaction between biofilms of Pseudomonas aeruginosa and PMNs in a murine implant model and in lung tissue from chronically infected cystic fibrosis patients. PNA FISH, DNA staining, labeling of PMN DNA with a thymidine analogue and immunohistochemistry were applied to localize bacteria, eDNA, PMN-derived eDNA, PMN-derived histone H3 (H3), neutrophil elastase (NE) and citrullinated H3 (citH3). Host-derived eDNA was observed surrounding bacterial biofilms but not within the biofilms. H3 localized to the lining of biofilms while NE was found throughout biofilms. CitH3, a marker for neutrophil extracellular traps (NETs) was detected only sporadically indicating that most host-derived eDNA in vivo was not a result of NETosis. Together these observations show that, in these in vivo biofilm infections with P. aeruginosa, the majority of eDNA is found external to the biofilm and derives from the host.
Keywords: NETosis; elastase; histone; neutrophil extracellular traps; polymorphonuclear leukocyte.
© FEMS 2020.
Figures





References
-
- Ahmed K, Dai TC, Ichinose Aet al. .. Neutrophil response to Pseudomonas aeruginosa in respiratory infection. Microbiol Immunol. 1993;37:523–9. - PubMed
-
- Alhede M, Bjarnsholt T, Jensen PØet al. .. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology. 2009;155:3500–8. - PubMed
-
- Allesen-Holm M, Barken KB, Yang Let al. .. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

