Drosophila-associated bacteria differentially shape the nutritional requirements of their host during juvenile growth
- PMID: 32196485
- PMCID: PMC7112240
- DOI: 10.1371/journal.pbio.3000681
Drosophila-associated bacteria differentially shape the nutritional requirements of their host during juvenile growth
Abstract
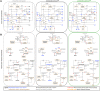
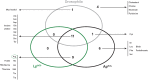
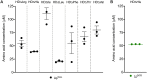
The interplay between nutrition and the microbial communities colonizing the gastrointestinal tract (i.e., gut microbiota) determines juvenile growth trajectory. Nutritional deficiencies trigger developmental delays, and an immature gut microbiota is a hallmark of pathologies related to childhood undernutrition. However, how host-associated bacteria modulate the impact of nutrition on juvenile growth remains elusive. Here, using gnotobiotic Drosophila melanogaster larvae independently associated with Acetobacter pomorumWJL (ApWJL) and Lactobacillus plantarumNC8 (LpNC8), 2 model Drosophila-associated bacteria, we performed a large-scale, systematic nutritional screen based on larval growth in 40 different and precisely controlled nutritional environments. We combined these results with genome-based metabolic network reconstruction to define the biosynthetic capacities of Drosophila germ-free (GF) larvae and its 2 bacterial partners. We first established that ApWJL and LpNC8 differentially fulfill the nutritional requirements of the ex-GF larvae and parsed such difference down to individual amino acids, vitamins, other micronutrients, and trace metals. We found that Drosophila-associated bacteria not only fortify the host's diet with essential nutrients but, in specific instances, functionally compensate for host auxotrophies by either providing a metabolic intermediate or nutrient derivative to the host or by uptaking, concentrating, and delivering contaminant traces of micronutrients. Our systematic work reveals that beyond the molecular dialogue engaged between the host and its bacterial partners, Drosophila and its associated bacteria establish an integrated nutritional network relying on nutrient provision and utilization.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Fairweather-Tait SJ, Southon S. Bioavailability of nutrients In: Caballero B, editor. Encyclopedia of Food Sciences and Nutrition (Second Edition). Oxford: Academic Press; 2003. pp. 478–484. 10.1016/B0-12-227055-X/00096-1 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

