Antibody characterization using immunosignatures
- PMID: 32196507
- PMCID: PMC7083272
- DOI: 10.1371/journal.pone.0229080
Antibody characterization using immunosignatures
Abstract
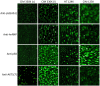
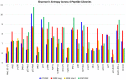
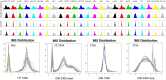
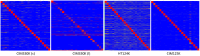
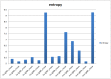
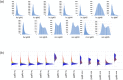
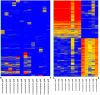
Therapeutic monoclonal antibodies have the potential to work as biological therapeutics. OKT3, Herceptin, Keytruda and others have positively impacted healthcare. Antibodies evolved naturally to provide high specificity and high affinity once mature. These characteristics can make them useful as therapeutics. However, we may be missing characteristics that are not obvious. We present a means of measuring antibodies in an unbiased manner that may highlight therapeutic activity. We propose using a microarray of random peptides to assess antibody properties. We tested twenty-four different commercial antibodies to gain some perspective about how much information can be derived from binding antibodies to random peptide libraries. Some monoclonals preferred to bind shorter peptides, some longer, some preferred motifs closer to the C-term, some nearer the N-term. We tested some antibodies with clinical activity but whose function was blinded to us at the time. We were provided with twenty-one different monoclonal antibodies, thirteen mouse and eight human IgM. These antibodies produced a variety of binding patterns on the random peptide arrays. When unblinded, the antibodies with polyspecific binding were the ones with the greatest therapeutic activity. The protein target to these therapeutic monoclonals is still unknown but using common sequence motifs from the peptides we predicted several human and mouse proteins. The same five highest proteins appeared in both mouse and human lists.
Conflict of interest statement
The authors have declared that no competing interest exist.
Figures

References
-
- Rodriguez M., et al. , Remyelination by Oligodendrocytes Stimulated by Antiserum to Spinal Cord. Journal of Neuropathology & Experimental Neurology, 1987. 46(1): p. 84–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

