Identification, Characterization and Expression Analysis of TRP Channel Genes in the Vegetable Pest, Pieris rapae
- PMID: 32197450
- PMCID: PMC7143563
- DOI: 10.3390/insects11030192
Identification, Characterization and Expression Analysis of TRP Channel Genes in the Vegetable Pest, Pieris rapae
Abstract
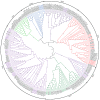
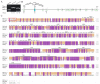
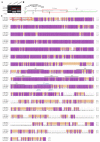
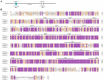
Transient receptor potential (TRP) channels are critical for insects to detect environmental stimuli and regulate homeostasis. Moreover, this superfamily has become potential molecular targets for insecticides or repellents. Pieris rapae is one of the most common and widely spread pests of Brassicaceae plants. Therefore, it is necessary to study TRP channels (TRPs) in P. rapae. In this study, we identified 14 TRPs in P. rapae, including two Water witch (Wtrw) genes. By contrast, only one Wtrw gene exists in Drosophila and functions in hygrosensation. We also found splice isoforms of Pyrexia (Pyx), TRPgamma (TRPγ) and TRP-Melastatin (TRPM). These three genes are related to temperature and gravity sensation, fine motor control, homeostasis regulation of Mg2+ and Zn2+ in Drosophila, respectively. Evolutionary analysis showed that the TRPs of P. rapae were well clustered into their own subfamilies. Real-time quantitative PCR (qPCR) showed that PrTRPs were widely distributed in the external sensory organs, including antennae, mouthparts, legs, wings and in the internal physiological organs, including brains, fat bodies, guts, Malpighian tubules, ovaries, as well as testis. Our study established a solid foundation for functional studies of TRP channels in P. rapae, and would be benefit to developing new approaches to control P. rapae targeting these important ion channels.
Keywords: Pieris rapae; gene duplication; splice isoform; transient receptor potential channel.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

