Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors
- PMID: 32198201
- PMCID: PMC7149396
- DOI: 10.1073/pnas.1921485117
Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors
Abstract
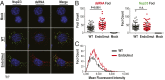
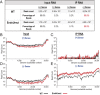
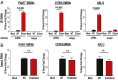
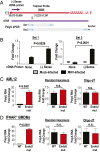
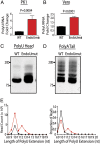
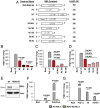
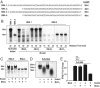
Coronaviruses (CoVs) are positive-sense RNA viruses that can emerge from endemic reservoirs and infect zoonotically, causing significant morbidity and mortality. CoVs encode an endoribonuclease designated EndoU that facilitates evasion of host pattern recognition receptor MDA5, but the target of EndoU activity was not known. Here, we report that EndoU cleaves the 5'-polyuridines from negative-sense viral RNA, termed PUN RNA, which is the product of polyA-templated RNA synthesis. Using a virus containing an EndoU catalytic-inactive mutation, we detected a higher abundance of PUN RNA in the cytoplasm compared to wild-type-infected cells. Furthermore, we found that transfecting PUN RNA into cells stimulates a robust, MDA5-dependent interferon response, and that removal of the polyuridine extension on the RNA dampens the response. Overall, the results of this study reveal the PUN RNA to be a CoV MDA5-dependent pathogen-associated molecular pattern (PAMP). We also establish a mechanism for EndoU activity to cleave and limit the accumulation of this PAMP. Since EndoU activity is highly conserved in all CoVs, inhibiting this activity may serve as an approach for therapeutic interventions against existing and emerging CoV infections.
Keywords: EndoU; coronavirus; endoribonuclease; interferon; nsp15.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Kato H., et al. , Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

